You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001332_00860
You are here: Home > Sequence: MGYG000001332_00860
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
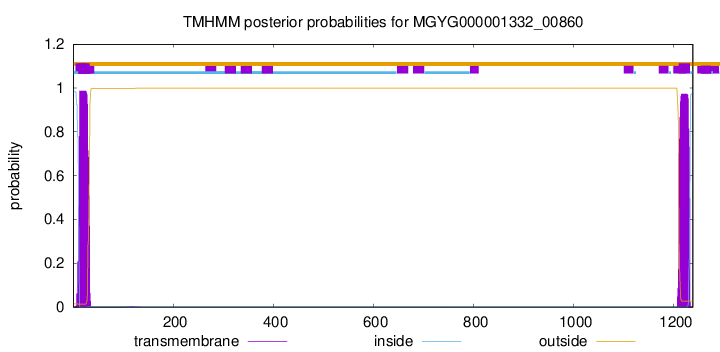
TMHMM annotations
Basic Information help
| Species | Massilioclostridium methylpentosum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Massilioclostridium; Massilioclostridium methylpentosum | |||||||||||
| CAZyme ID | MGYG000001332_00860 | |||||||||||
| CAZy Family | cohesin | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1018339; End: 1022055 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| cohesin | 39 | 158 | 7.1e-17 | 0.9253731343283582 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd08548 | Type_I_cohesin_like | 1.45e-16 | 39 | 171 | 1 | 135 | Type I cohesin domain, interaction partner of dockerin. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I cohesins; their interactions with dockerin mediate assembly of a range of dockerin-borne enzymes to the complex. |
| COG5492 | YjdB | 2.55e-14 | 223 | 407 | 143 | 321 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| COG5492 | YjdB | 3.67e-14 | 204 | 494 | 27 | 326 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| COG5492 | YjdB | 7.76e-14 | 785 | 992 | 128 | 321 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| COG5492 | YjdB | 2.07e-11 | 367 | 649 | 33 | 317 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANQ52856.2 | 1.53e-49 | 256 | 1011 | 712 | 1551 |
| QWG05121.1 | 3.12e-48 | 256 | 1017 | 712 | 1555 |
| QWG05123.1 | 2.40e-41 | 256 | 1003 | 703 | 1538 |
| BCT44110.1 | 9.35e-39 | 261 | 824 | 318 | 826 |
| ATU19782.1 | 6.70e-35 | 249 | 651 | 955 | 1399 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q086E4 | 1.02e-29 | 257 | 926 | 40 | 653 | Ice-binding protein 1 OS=Shewanella frigidimarina (strain NCIMB 400) OX=318167 GN=Sfri_1018 PE=1 SV=1 |
| P33747 | 2.07e-13 | 764 | 913 | 39 | 189 | Uncharacterized protein CA_P0160 OS=Clostridium acetobutylicum (strain ATCC 824 / DSM 792 / JCM 1419 / LMG 5710 / VKM B-1787) OX=272562 GN=CA_P0160 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
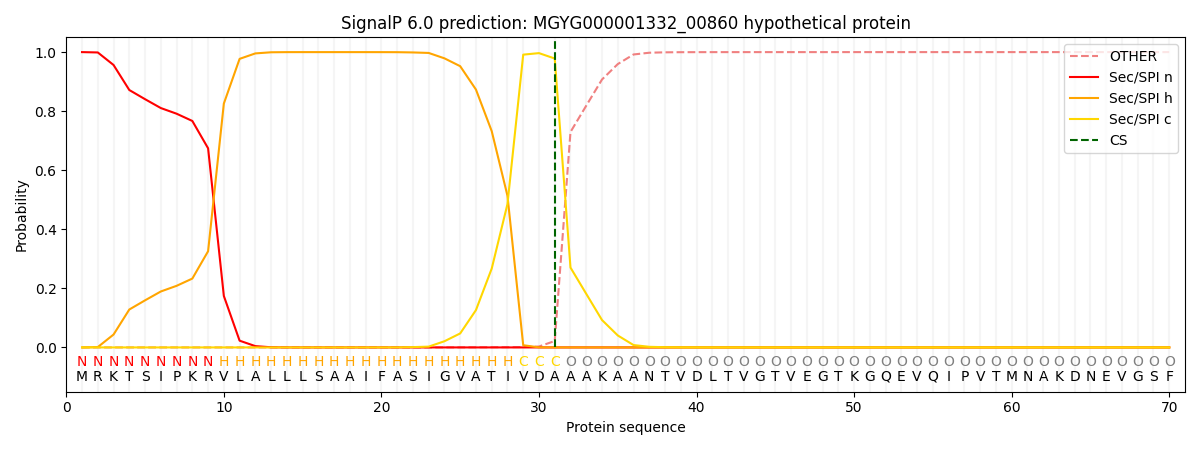
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000284 | 0.999027 | 0.000172 | 0.000184 | 0.000161 | 0.000140 |