You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001332_01065
You are here: Home > Sequence: MGYG000001332_01065
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
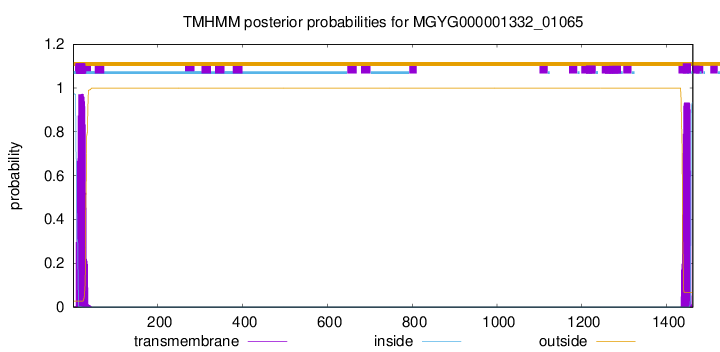
TMHMM annotations
Basic Information help
| Species | Massilioclostridium methylpentosum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Massilioclostridium; Massilioclostridium methylpentosum | |||||||||||
| CAZyme ID | MGYG000001332_01065 | |||||||||||
| CAZy Family | CBM67 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1278633; End: 1283021 Strand: + | |||||||||||
Full Sequence Download help
| MKTSAKAKII SVVLSIAMIA SVATQAFAAV PSGTAASTAI VSLKTNNHVD PLNVDDTTPT | 60 |
| FSWQMQSNVI GAMQSAYQIV VTDPNGSTVW DSGKQYDSAS AAVEYAGDAL SPTTTYSWTV | 120 |
| TVWDTAGNSY TSEPASFETG FMDTTIDAWD GAQWIGTQER NLDATSLYIF NMGYKLRLVP | 180 |
| ETTKASFIFG ADDFRLTNKY LSELLTGTEN YIRVELDITD MTEEGGAKLN IYRPGYSQTN | 240 |
| ADPDVPYLTI SAESGSNIDE ILTYANRYDT HSFDFTVNAS SISLKIDGTQ LQLSGGSRPT | 300 |
| TSFKVKDGET GNNYNTYPDL NQIGFQADPG QVAVFSDLYV ANFDLWASDK VGARLFDSET | 360 |
| GATYSIFDGI KGVSHTGNEL FINGGLTGVF GYADPSYGSV PMLRTEINAS KEIADARLFV | 420 |
| TAHGIYEMYI NGERVGEDYF NPGVSEYEKN LRYTAYDVTS MLNQGVNAVG ALLGPGWWSD | 480 |
| SMSYTPDNYN YYGDQQALLA KIVVKYTDGT SDTFVTNPDT WTTSNESPIR YSSFFQGERY | 540 |
| DATMEAAVEG WNTAGFDASD WVNASVIETR EKFANAALTG DENDPGRVAE VVDSTYVGES | 600 |
| KEGSGSYIYD MGTNMLGIPQ ITIPEGYASE GQEVIIRYAE VFYPELDEYV EQGVDGLLMV | 660 |
| ENYRAALSTD FYTCKDGAQT IQPTFTYHGY RYLEITGLDK PLPAECVKSL SLSSIEEMTG | 720 |
| SYNSSNELVN QLFKNVQRSQ YSNFLSLPTD CPQRNERMGW LGDAQVFART ASYNADVSNI | 780 |
| YQSFLDIIRD DQTEEGHISV YAPSYGGSGF WSGTSWQSAI AIIPYQMYMQ TGNTDIIREN | 840 |
| IDAMYSYLGY LAAAPLSEAA PHLTSRAGFL ADWLSIDSTD SNLINNAVYV YTMDIFVQMA | 900 |
| NVIGDERAAE LAERYEAAKA EWNALYIDPE TGRTTDHTQA SYATPLMYGV INDEYVEKAA | 960 |
| ANLVDTVENP QNPEVKPYTI TTGFSGTPFI VPSLTNYGYV DDAYKMLEQT EYASWLYPVT | 1020 |
| QGATSIWERW NSYTLEGGFN GNNSMNSFNH YSLGAIAEWM YGYQLGITNA EGVAAYKEFV | 1080 |
| LQPTVGGSFT FANGSFDSKY GTIESGWTAA DGAMDSYSAT VPANTTATLY LPVSEEATAN | 1140 |
| FADVDGITYL GRTTHNNQSV AAFKLVAGEY SFNLVDGKLV ASVADGYVSG PDIGKDILRK | 1200 |
| TIAYAQEQKA DPSFENVIPM VQESFNAALE NAIAVEANPA ADKDMVYNAW TTLMNEIHKL | 1260 |
| GFVKGDKTNL EKLILAGDGI LVDIDNYIED GKQEFTDALA AAKVVFNDPN ALEGEVTEAS | 1320 |
| DRLLDAMVNL RLKADKSLLG KTIEVANSID TAVYTLESVN VFNTAKAEAE AIYNDDAYDR | 1380 |
| DQEAINAATN KLQNAIDGLV PIDSSTDLDA NTSTPDVPVQ GDATATTGTS TPKTGETAPA | 1440 |
| AIAVAMLALA GAAAVLGKKN RK | 1462 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH78 | 601 | 1131 | 2.8e-147 | 0.9940476190476191 |
| CBM67 | 397 | 568 | 2.7e-31 | 0.8863636363636364 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17389 | Bac_rhamnosid6H | 4.25e-102 | 717 | 1063 | 1 | 339 | Bacterial alpha-L-rhamnosidase 6 hairpin glycosidase domain. This family consists of bacterial rhamnosidase A and B enzymes. L-Rhamnose is abundant in biomass as a common constituent of glycolipids and glycosides, such as plant pigments, pectic polysaccharides, gums or biosurfactants. Some rhamnosides are important bioactive compounds. For example, terpenyl glycosides, the glycosidic precursor of aromatic terpenoids, act as important flavouring substances in grapes. Other rhamnosides act as cytotoxic rhamnosylated terpenoids, as signal substances in plants or play a role in the antigenicity of pathogenic bacteria. |
| pfam08531 | Bac_rhamnosid_N | 1.49e-49 | 411 | 568 | 1 | 154 | Alpha-L-rhamnosidase N-terminal domain. This family consists of bacterial rhamnosidase A and B enzymes. This domain is probably involved in substrate recognition. |
| pfam05592 | Bac_rhamnosid | 4.13e-20 | 601 | 703 | 2 | 93 | Bacterial alpha-L-rhamnosidase concanavalin-like domain. This family consists of bacterial rhamnosidase A and B enzymes. L-Rhamnose is abundant in biomass as a common constituent of glycolipids and glycosides, such as plant pigments, pectic polysaccharides, gums or biosurfactants. Some rhamnosides are important bioactive compounds. For example, terpenyl glycosides, the glycosidic precursor of aromatic terpenoids, act as important flavouring substances in grapes. Other rhamnosides act as cytotoxic rhamnosylated terpenoids, as signal substances in plants or play a role in the antigenicity of pathogenic bacteria. |
| pfam17390 | Bac_rhamnosid_C | 3.04e-18 | 1071 | 1140 | 4 | 72 | Bacterial alpha-L-rhamnosidase C-terminal domain. This family consists of bacterial rhamnosidase A and B enzymes. L-Rhamnose is abundant in biomass as a common constituent of glycolipids and glycosides, such as plant pigments, pectic polysaccharides, gums or biosurfactants. Some rhamnosides are important bioactive compounds. For example, terpenyl glycosides, the glycosidic precursor of aromatic terpenoids, act as important flavouring substances in grapes. Other rhamnosides act as cytotoxic rhamnosylated terpenoids, as signal substances in plants or play a role in the antigenicity of pathogenic bacteria. |
| pfam07554 | FIVAR | 0.006 | 1354 | 1400 | 22 | 69 | FIVAR domain. This domain is found in a wide variety of contexts, but mostly occurring in cell wall associated proteins. A lack of conserved catalytic residues suggests that it is a binding domain. From context, possible substrates are hyaluronate or fibronectin (personal obs: C Yeats). This is further evidenced by. Possibly the exact substrate is N-acetyl glucosamine. Finding it in the same protein as pfam05089 further supports this proposal. It is found in the C-terminal part of Bacillus sp. Gellan lyase, which is removed during maturation. Some of the proteins it is found in are involved in methicillin resistance. The name FIVAR derives from Found In Various Architectures. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGH33563.1 | 7.65e-306 | 37 | 1193 | 39 | 1210 |
| AHF25035.1 | 4.01e-280 | 36 | 1179 | 19 | 1231 |
| QGH37059.1 | 2.10e-275 | 40 | 1175 | 32 | 1183 |
| AOZ98905.1 | 3.42e-269 | 27 | 1175 | 11 | 1180 |
| AIF65800.1 | 2.63e-262 | 40 | 1175 | 25 | 1162 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3W5M_A | 1.65e-134 | 51 | 1183 | 17 | 1036 | CrystalStructure of Streptomyces avermitilis alpha-L-rhamnosidase [Streptomyces avermitilis MA-4680 = NBRC 14893],3W5N_A Crystal Structure of Streptomyces avermitilis alpha-L-rhamnosidase complexed with L-rhamnose [Streptomyces avermitilis MA-4680 = NBRC 14893] |
| 6GSZ_A | 4.46e-96 | 404 | 1132 | 135 | 847 | Crystalstructure of native alfa-L-rhamnosidase from Aspergillus terreus [Aspergillus terreus] |
| 6I60_A | 1.41e-91 | 401 | 1172 | 177 | 939 | Structureof alpha-L-rhamnosidase from Dictyoglumus thermophilum [Dictyoglomus thermophilum H-6-12],6I60_B Structure of alpha-L-rhamnosidase from Dictyoglumus thermophilum [Dictyoglomus thermophilum H-6-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q82PP4 | 9.26e-134 | 51 | 1132 | 17 | 995 | Alpha-L-rhamnosidase OS=Streptomyces avermitilis (strain ATCC 31267 / DSM 46492 / JCM 5070 / NBRC 14893 / NCIMB 12804 / NRRL 8165 / MA-4680) OX=227882 GN=SAVERM_828 PE=1 SV=1 |
| T2KNB2 | 8.12e-114 | 388 | 1132 | 162 | 879 | Alpha-L-rhamnosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22090 PE=1 SV=2 |
| P9WF03 | 1.35e-109 | 393 | 1175 | 168 | 911 | Alpha-L-rhamnosidase OS=Alteromonas sp. (strain LOR) OX=1537994 GN=LOR_34 PE=1 SV=1 |
| T2KPL4 | 2.55e-62 | 404 | 1158 | 188 | 925 | Alpha-L-rhamnosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22170 PE=2 SV=1 |
| T2KM26 | 1.24e-07 | 690 | 1032 | 700 | 1048 | Bifunctional sulfatase/alpha-L-rhamnosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22250 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
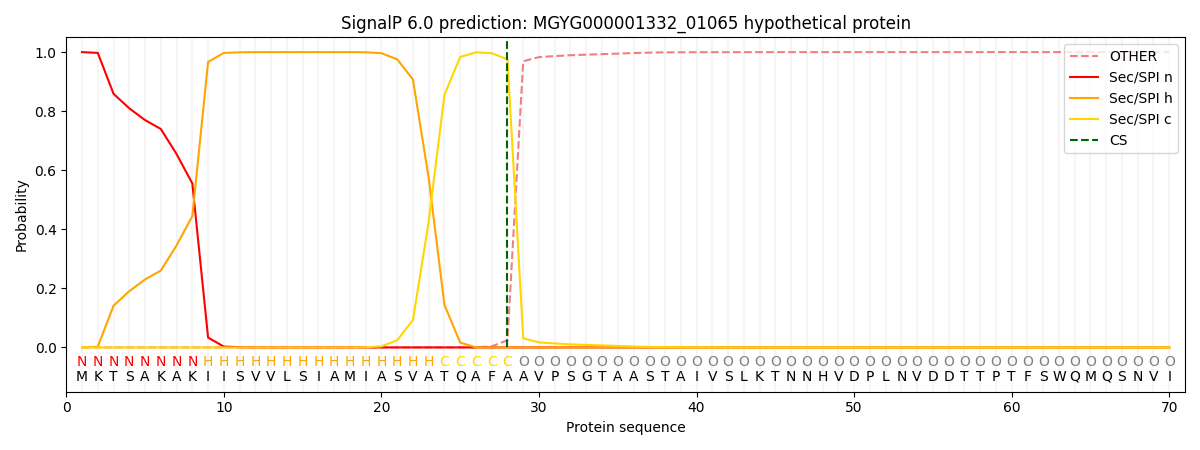
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000320 | 0.998876 | 0.000177 | 0.000235 | 0.000189 | 0.000166 |