You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001332_01105
You are here: Home > Sequence: MGYG000001332_01105
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
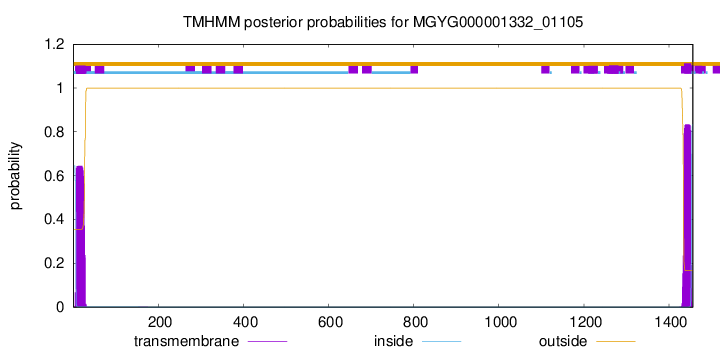
TMHMM annotations
Basic Information help
| Species | Massilioclostridium methylpentosum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Massilioclostridium; Massilioclostridium methylpentosum | |||||||||||
| CAZyme ID | MGYG000001332_01105 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1337387; End: 1341757 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 492 | 1069 | 1.1e-61 | 0.6820388349514563 |
| CBM32 | 350 | 464 | 3.3e-17 | 0.9274193548387096 |
| CBM32 | 204 | 326 | 1.6e-16 | 0.9435483870967742 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 5.81e-27 | 483 | 1027 | 362 | 872 | alpha-L-rhamnosidase. |
| pfam00754 | F5_F8_type_C | 7.02e-14 | 203 | 325 | 2 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 5.24e-12 | 349 | 464 | 3 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| COG5281 | COG5281 | 2.33e-06 | 1080 | 1396 | 233 | 565 | Phage-related minor tail protein [Mobilome: prophages, transposons]. |
| COG4372 | COG4372 | 3.04e-06 | 1257 | 1424 | 53 | 235 | Uncharacterized conserved protein, contains DUF3084 domain [Function unknown]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCI61176.1 | 0.0 | 1 | 1072 | 1 | 1059 |
| QGT72311.1 | 7.15e-126 | 484 | 1049 | 183 | 723 |
| QGA23530.1 | 2.21e-123 | 488 | 1049 | 186 | 730 |
| BCG54873.1 | 3.63e-123 | 482 | 1050 | 181 | 750 |
| AWW28862.1 | 4.82e-117 | 449 | 1050 | 157 | 754 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q2F_A | 3.37e-09 | 571 | 1070 | 512 | 993 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
| 2RV9_A | 4.62e-08 | 349 | 467 | 15 | 135 | Solutionstructure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 4ZXE_A | 4.72e-08 | 349 | 467 | 16 | 136 | X-raycrystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_B X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_C X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis] |
| 4ZY9_A | 4.72e-08 | 349 | 467 | 16 | 136 | X-raycrystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZY9_B X-ray crystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 5MQM_A | 3.79e-06 | 686 | 1048 | 580 | 919 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KNA8 | 4.78e-09 | 571 | 1071 | 310 | 757 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
| E8MGH9 | 1.18e-06 | 1057 | 1272 | 1666 | 1885 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
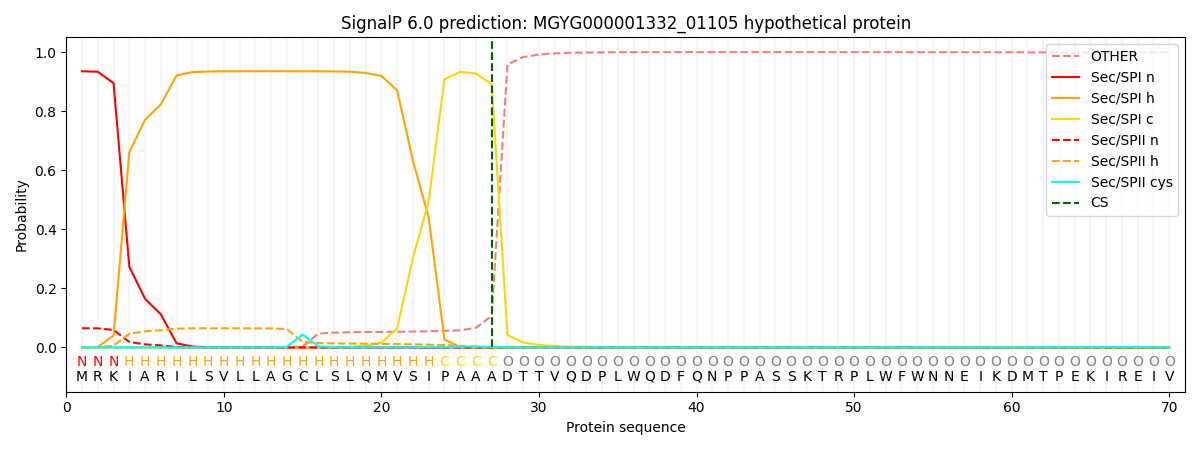
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000627 | 0.923943 | 0.074405 | 0.000381 | 0.000337 | 0.000261 |