You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001332_01308
You are here: Home > Sequence: MGYG000001332_01308
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
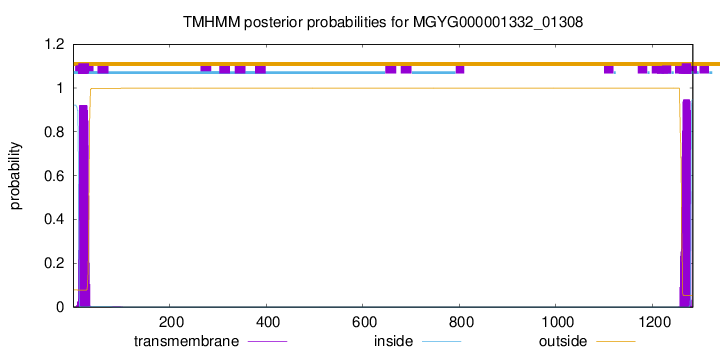
TMHMM annotations
Basic Information help
| Species | Massilioclostridium methylpentosum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Massilioclostridium; Massilioclostridium methylpentosum | |||||||||||
| CAZyme ID | MGYG000001332_01308 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 54339; End: 58193 Strand: - | |||||||||||
Full Sequence Download help
| MKSANWWKKC LAALLSAALI SSCVGSAGVF AAGDASTGEP TLAEEFQNAP QQSRTMIRYW | 60 |
| LPDAAVEEEE VRRDIQTIAE LGYGGIEVMG LTMLTSGVTD DYRWGTENWD RAMEVVVDEA | 120 |
| KKQGLTVDVT NGPGWPISMP GIETADDEAA SYEMTYGTAI VAGGERYTGV VPARNQVRKS | 180 |
| GTTKLFAVAA YRVAGDKTVD YSSYIDLMPS VALNEQDPAQ STLDWTAPDG EDWMVFSFWE | 240 |
| QPTAQQYGGK YYIVDHFGRA GAQACIDYWE QVLEEKNYLH EVSSIFNDSL EYQVDKEWTR | 300 |
| GFQEIFEQQK DYDITPYLPV IGGTGYYQTG EGPNFSFTDS ELTQRIKNDY NDVVTYCYNE | 360 |
| YHLKPLQQMA EKHGMNIRYQ VAYNKPMEVE TSALSVGIPE GEGLNRATLD NLRMMASAVH | 420 |
| MTDKQIYSFE AAAEFGNANG QSYEDMAWWL KRSWAAGMNR QVLHGASYSG VWEDGTLNGR | 480 |
| FGGTSWPGWS GFSGFISNDW NRNTSEENAG SYIEYFSRYN YIMQKESKID VAVYRHTYED | 540 |
| RYMLSGNDGE DWYPDGGALN ANGYSYEFVS PSILSLDTAK VTNGVLNENG PAYKAIVLAN | 600 |
| EERMSPSTLD TLTEMVEAGL KLVVVGDTPS KLMYQSELNE GYTDAMIVDG MASLCENGNV | 660 |
| RQVADYADVP AALAALGVTA DAQYSTPTDI LARHQVDEIG DYYYLYNYNK MLAEDANATL | 720 |
| ATAGTGYPGI NKEMAFSEKD VEVTLKGEGR PYLLDGWTGE ITPIASYQLN NGSVTLKLSF | 780 |
| DEDEAKLIAL LTDEEAQANG ITPRSSYVTA LDSSSQAVYT DQGEIALRAS EEGRHTATLS | 840 |
| NGTQCAASVE EVQQSFTVGA WDLQISAIEP GEGSIYFRDS VWNQLEPTTV TSLAGWSEIN | 900 |
| PDWKNVSGIG RYTATFTLDK GWEQGYGATI ALGEVEDTYT VEVNGTQLPF TNQINTTLDI | 960 |
| GPYVHQGENT IVVTVASTLY NQFKAHGGMM SSMPLKQNGL LGVNGVVRVT PYQQVVFEAD | 1020 |
| TSKEILNKVL AYAQQQYEDP SFEQVIAVVQ ETFTAALESA RAVAADPYAT QQEIDAAWQT | 1080 |
| LMTEIHKLGF VRGDKTSLAT LLEVATQYEV KIDSYTPTTA DQFVAVLAEA RETYADENAM | 1140 |
| QDDVSAAESD LLEAMMNLRY KADKGVLESV LAKASEIDLA GYTAESRAAF QTANDNARAV | 1200 |
| YEDENATQDE VNSAADRLHA AIDRLEATDA ASGSIVGEGA SIQGDATRTN GSATAKTGET | 1260 |
| TPVAALVAVL ALAGAGLIVS KKKR | 1284 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 43 | 797 | 5.6e-81 | 0.787621359223301 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 2.10e-24 | 193 | 707 | 342 | 837 | alpha-L-rhamnosidase. |
| pfam02837 | Glyco_hydro_2_N | 6.49e-05 | 884 | 975 | 41 | 133 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| pfam07554 | FIVAR | 8.00e-04 | 1167 | 1226 | 3 | 69 | FIVAR domain. This domain is found in a wide variety of contexts, but mostly occurring in cell wall associated proteins. A lack of conserved catalytic residues suggests that it is a binding domain. From context, possible substrates are hyaluronate or fibronectin (personal obs: C Yeats). This is further evidenced by. Possibly the exact substrate is N-acetyl glucosamine. Finding it in the same protein as pfam05089 further supports this proposal. It is found in the C-terminal part of Bacillus sp. Gellan lyase, which is removed during maturation. Some of the proteins it is found in are involved in methicillin resistance. The name FIVAR derives from Found In Various Architectures. |
| NF033914 | antiphage_ZorA_1 | 0.006 | 1121 | 1257 | 351 | 501 | anti-phage defense protein ZorA. Proteins of this subfamily are putative H+ channel proteins, but it has been reported that they are also involved in anti-phage defense. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AND79497.1 | 1.97e-143 | 42 | 1018 | 36 | 1002 |
| ALC24789.1 | 1.95e-127 | 43 | 1022 | 62 | 1018 |
| QMU12900.1 | 1.95e-127 | 43 | 1022 | 62 | 1018 |
| QFY07470.1 | 4.02e-126 | 54 | 1012 | 47 | 965 |
| QKW37235.1 | 2.46e-121 | 54 | 1019 | 60 | 984 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
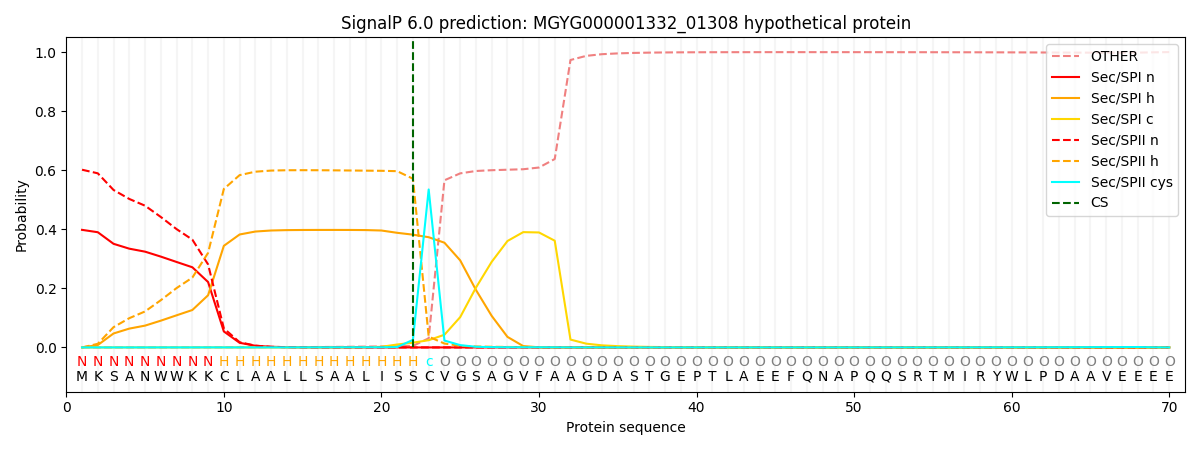
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000595 | 0.390637 | 0.608013 | 0.000312 | 0.000233 | 0.000183 |