You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001332_01584
You are here: Home > Sequence: MGYG000001332_01584
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
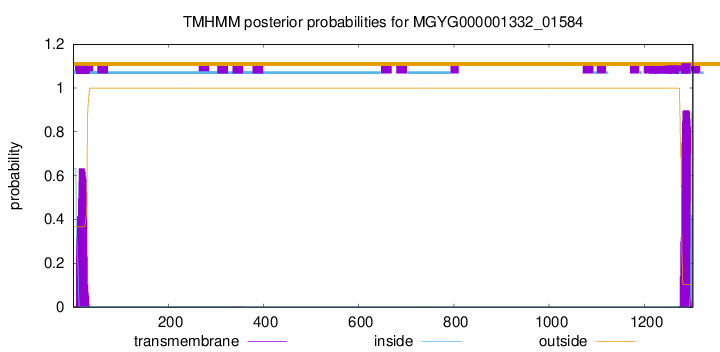
TMHMM annotations
Basic Information help
| Species | Massilioclostridium methylpentosum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Massilioclostridium; Massilioclostridium methylpentosum | |||||||||||
| CAZyme ID | MGYG000001332_01584 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1524; End: 5432 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 44 | 828 | 3.5e-74 | 0.8143203883495146 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 1.90e-13 | 171 | 776 | 313 | 869 | alpha-L-rhamnosidase. |
| pfam02837 | Glyco_hydro_2_N | 2.96e-06 | 919 | 991 | 61 | 133 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| PRK10340 | ebgA | 0.001 | 925 | 991 | 111 | 177 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| TIGR01167 | LPXTG_anchor | 0.007 | 1274 | 1302 | 4 | 32 | LPXTG-motif cell wall anchor domain. This model describes the LPXTG motif-containing region found at the C-terminus of many surface proteins of Streptococcus and Streptomyces species. Cleavage between the Thr and Gly by sortase or a related enzyme leads to covalent anchoring at the new C-terminal Thr to the cell wall. Hits that do not lie at the C-terminus or are not found in Gram-positive bacteria are probably false-positive. A common feature of this proteins containing this domain appears to be a high proportion of charged and zwitterionic residues immediatedly upstream of the LPXTG motif. This model differs from other descriptions of the LPXTG region by including a portion of that upstream charged region. [Cell envelope, Other] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGH70002.1 | 7.19e-231 | 2 | 1044 | 43 | 1127 |
| QYN20448.1 | 1.75e-208 | 18 | 1033 | 23 | 1115 |
| SDG73971.1 | 6.37e-194 | 40 | 1031 | 3 | 955 |
| SDS67158.1 | 1.83e-191 | 40 | 1004 | 3 | 944 |
| QHC72677.1 | 2.95e-191 | 50 | 1004 | 14 | 938 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q2F_A | 1.96e-10 | 200 | 1000 | 402 | 1103 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
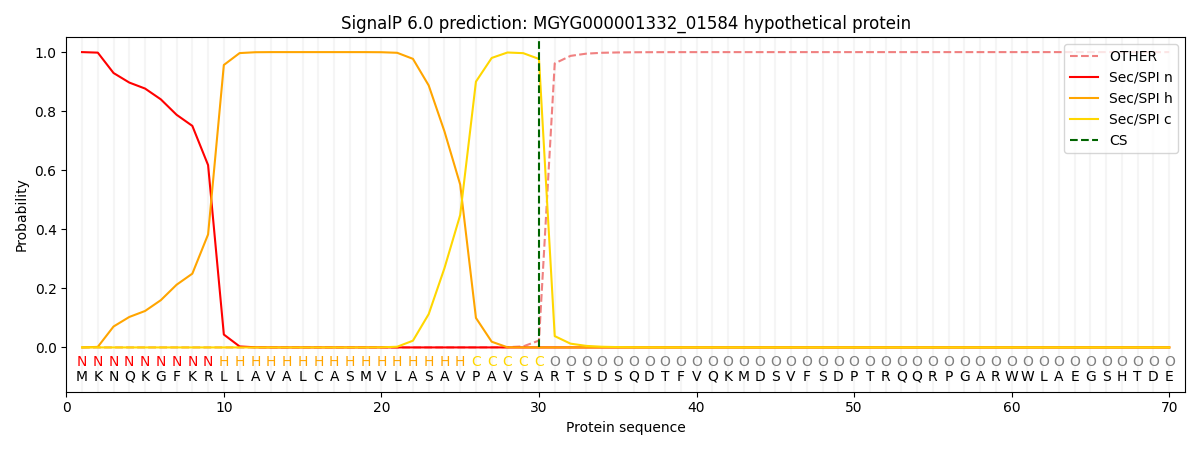
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000376 | 0.998783 | 0.000204 | 0.000229 | 0.000208 | 0.000180 |