You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001332_01592
You are here: Home > Sequence: MGYG000001332_01592
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
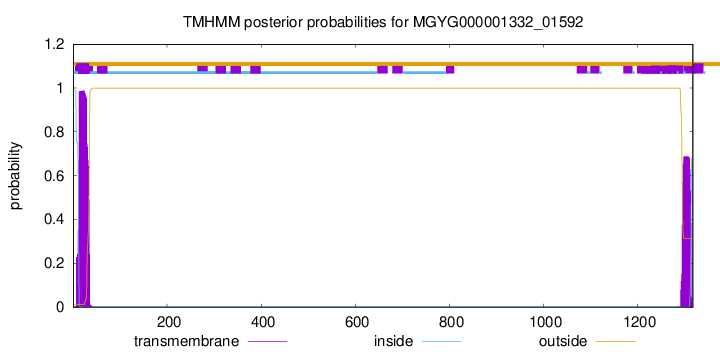
TMHMM annotations
Basic Information help
| Species | Massilioclostridium methylpentosum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Massilioclostridium; Massilioclostridium methylpentosum | |||||||||||
| CAZyme ID | MGYG000001332_01592 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 24877; End: 28833 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 46 | 970 | 1.6e-124 | 0.9599514563106796 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 3.47e-44 | 171 | 751 | 296 | 872 | alpha-L-rhamnosidase. |
| NF033930 | pneumo_PspA | 5.37e-05 | 997 | 1270 | 96 | 355 | pneumococcal surface protein A. The pneumococcal surface protein proteins, found in Streptococcus pneumoniae, are repetitive, with patterns of localized high sequence identity across pairs of proteins given different specific names that recombination may be presumed. This protein, PspA, has an N-terminal region that lacks a cross-wall-targeting YSIRK type extended signal peptide, in contrast to the closely related choline-binding protein CbpA which has a similar C-terminus but a YSIRK-containing region at the N-terminus. |
| COG4942 | EnvC | 5.97e-05 | 1044 | 1298 | 60 | 303 | Septal ring factor EnvC, activator of murein hydrolases AmiA and AmiB [Cell cycle control, cell division, chromosome partitioning]. |
| COG4372 | COG4372 | 1.02e-04 | 1058 | 1272 | 68 | 248 | Uncharacterized conserved protein, contains DUF3084 domain [Function unknown]. |
| TIGR02168 | SMC_prok_B | 2.92e-04 | 994 | 1272 | 728 | 980 | chromosome segregation protein SMC, common bacterial type. SMC (structural maintenance of chromosomes) proteins bind DNA and act in organizing and segregating chromosomes for partition. SMC proteins are found in bacteria, archaea, and eukaryotes. This family represents the SMC protein of most bacteria. The smc gene is often associated with scpB (TIGR00281) and scpA genes, where scp stands for segregation and condensation protein. SMC was shown (in Caulobacter crescentus) to be induced early in S phase but present and bound to DNA throughout the cell cycle. [Cellular processes, Cell division, DNA metabolism, Chromosome-associated proteins] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIY75684.1 | 1.43e-176 | 49 | 993 | 48 | 996 |
| QMV04309.1 | 1.27e-175 | 40 | 991 | 43 | 1000 |
| AXG51232.1 | 1.27e-175 | 40 | 991 | 43 | 1000 |
| ANS62302.1 | 1.27e-175 | 40 | 991 | 43 | 1000 |
| QDO03140.1 | 1.51e-175 | 49 | 993 | 48 | 996 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q2F_A | 3.62e-25 | 223 | 971 | 422 | 1109 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
| 5MQM_A | 3.37e-10 | 269 | 566 | 421 | 700 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A | 7.60e-10 | 269 | 566 | 421 | 700 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KNA8 | 1.45e-28 | 49 | 724 | 34 | 689 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
| E8MGH9 | 2.76e-09 | 1072 | 1318 | 1667 | 1932 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000704 | 0.998398 | 0.000205 | 0.000302 | 0.000194 | 0.000176 |