You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001332_02073
You are here: Home > Sequence: MGYG000001332_02073
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
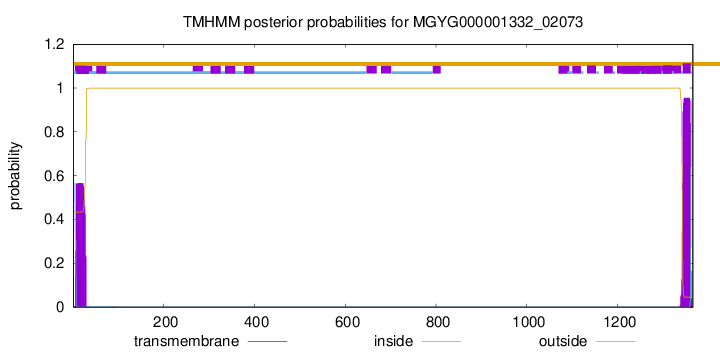
TMHMM annotations
Basic Information help
| Species | Massilioclostridium methylpentosum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Massilioclostridium; Massilioclostridium methylpentosum | |||||||||||
| CAZyme ID | MGYG000001332_02073 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 393451; End: 397551 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 39 | 758 | 4e-55 | 0.6407766990291263 |
| CBM32 | 775 | 892 | 1.4e-17 | 0.9354838709677419 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 2.43e-35 | 44 | 732 | 1 | 755 | alpha-L-rhamnosidase. |
| pfam00754 | F5_F8_type_C | 2.81e-13 | 205 | 321 | 4 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 2.81e-10 | 774 | 878 | 3 | 112 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 1.36e-06 | 205 | 324 | 16 | 142 | Substituted updates: Jan 31, 2002 |
| COG5283 | COG5283 | 0.001 | 1046 | 1333 | 66 | 317 | Phage-related tail protein [Mobilome: prophages, transposons]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCI61176.1 | 1.01e-211 | 36 | 1044 | 31 | 1055 |
| QGA23530.1 | 5.40e-140 | 41 | 1031 | 23 | 732 |
| BCG54873.1 | 1.96e-132 | 36 | 770 | 18 | 616 |
| QGT72311.1 | 7.10e-130 | 43 | 750 | 27 | 594 |
| AWW28862.1 | 3.03e-109 | 43 | 1030 | 35 | 754 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
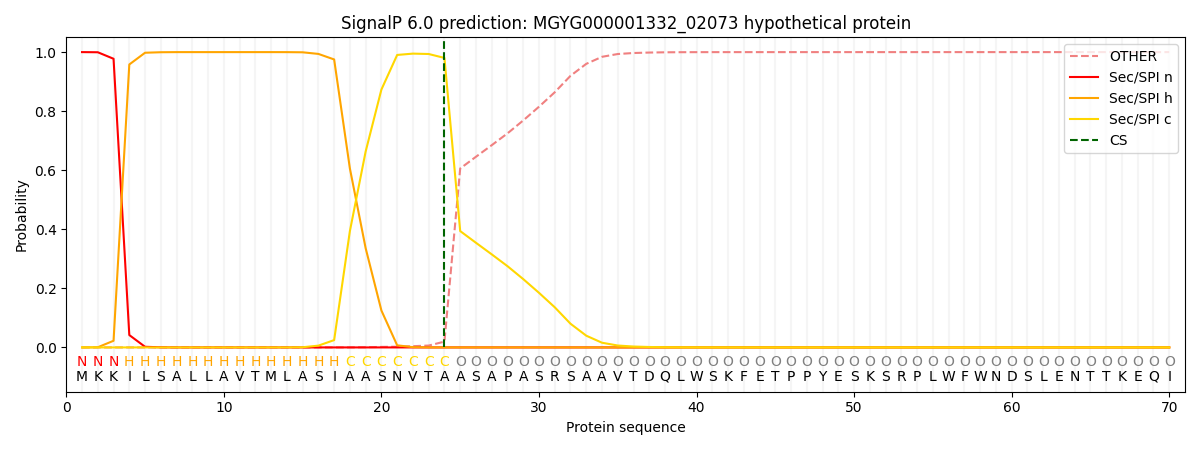
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000292 | 0.999037 | 0.000170 | 0.000179 | 0.000155 | 0.000141 |