You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001332_02312
Basic Information
help
| Species |
Massilioclostridium methylpentosum
|
| Lineage |
Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Massilioclostridium; Massilioclostridium methylpentosum
|
| CAZyme ID |
MGYG000001332_02312
|
| CAZy Family |
GH106 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 1122 |
|
122264.68 |
4.0251 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001332 |
3406391 |
Isolate |
not provided |
not provided |
|
| Gene Location |
Start: 94348;
End: 97716
Strand: -
|
No EC number prediction in MGYG000001332_02312.
| Family |
Start |
End |
Evalue |
family coverage |
| GH106 |
47 |
844 |
5.2e-99 |
0.8033980582524272 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam17132
|
Glyco_hydro_106 |
1.88e-32 |
259 |
795 |
365 |
872 |
alpha-L-rhamnosidase. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 6Q2F_A
|
2.01e-18 |
330 |
1039 |
499 |
1138 |
Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
| 5MQM_A
|
3.09e-13 |
334 |
598 |
458 |
687 |
Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A
|
5.31e-13 |
349 |
598 |
467 |
687 |
Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
T2KNA8
|
9.33e-21 |
298 |
821 |
264 |
742 |
Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
This protein is predicted as SP
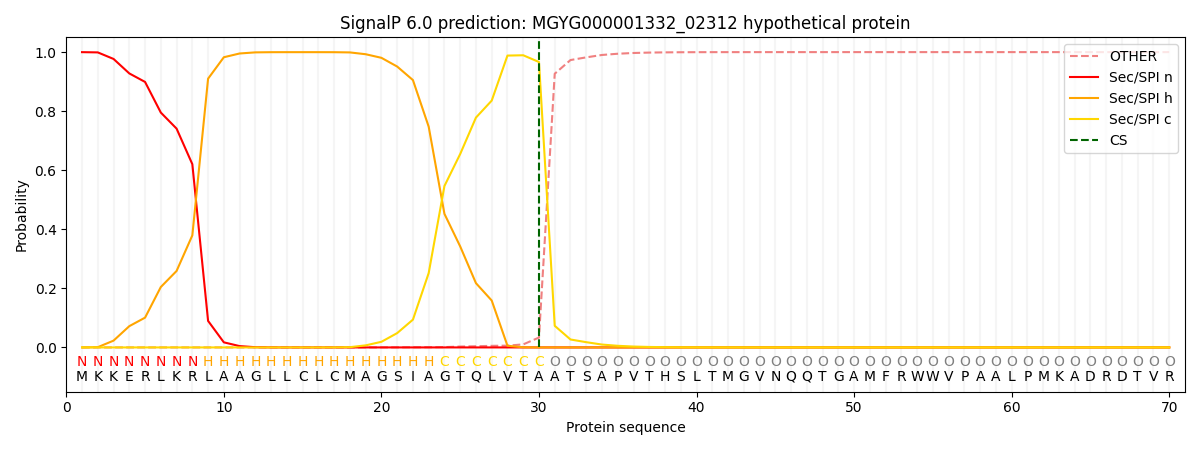
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000715
|
0.998058
|
0.000554
|
0.000253
|
0.000203
|
0.000183
|
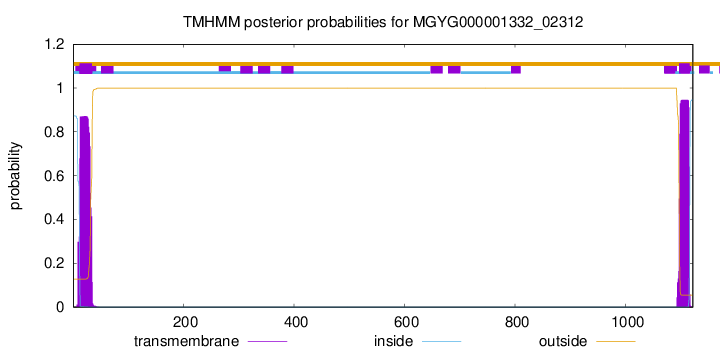
| start |
end |
| 12 |
34 |
| 1097 |
1116 |


