You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001332_02619
Basic Information
help
| Species |
Massilioclostridium methylpentosum
|
| Lineage |
Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Massilioclostridium; Massilioclostridium methylpentosum
|
| CAZyme ID |
MGYG000001332_02619
|
| CAZy Family |
GH106 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001332 |
3406391 |
Isolate |
not provided |
not provided |
|
| Gene Location |
Start: 444764;
End: 447964
Strand: +
|
No EC number prediction in MGYG000001332_02619.
| Family |
Start |
End |
Evalue |
family coverage |
| GH106 |
38 |
974 |
6e-101 |
0.9490291262135923 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam17132
|
Glyco_hydro_106 |
1.69e-24 |
324 |
762 |
448 |
872 |
alpha-L-rhamnosidase. |
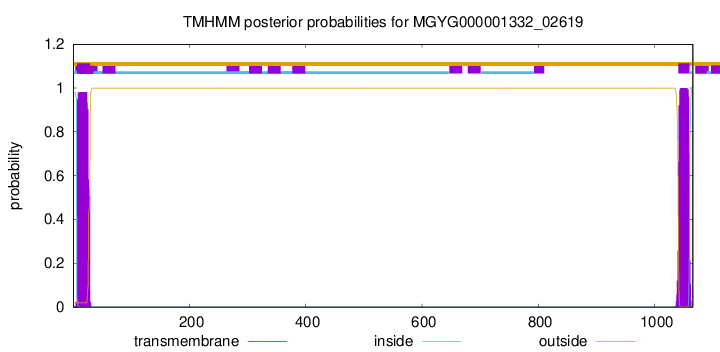
This protein is predicted as SP
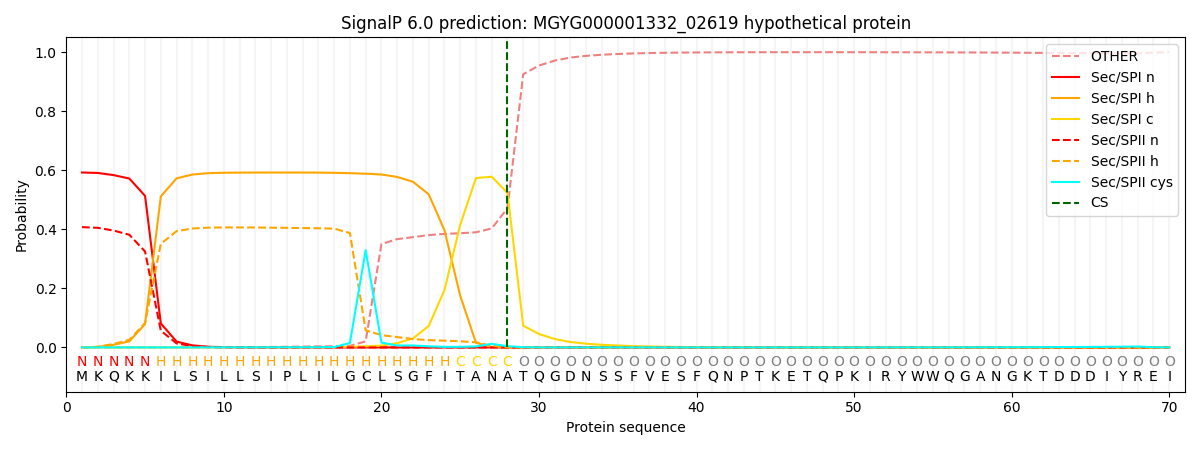
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.001260
|
0.578018
|
0.419413
|
0.000592
|
0.000378
|
0.000315
|