You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001334_01057
You are here: Home > Sequence: MGYG000001334_01057
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
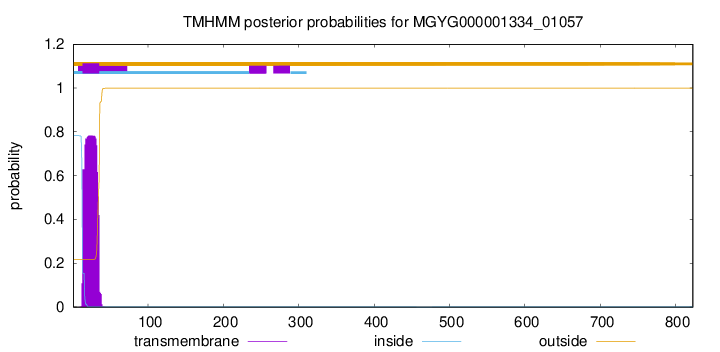
TMHMM annotations
Basic Information help
| Species | Ligilactobacillus ruminis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Ligilactobacillus; Ligilactobacillus ruminis | |||||||||||
| CAZyme ID | MGYG000001334_01057 | |||||||||||
| CAZy Family | GH25 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1067525; End: 1069996 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH25 | 97 | 279 | 4.5e-32 | 0.9830508474576272 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06522 | GH25_AtlA-like | 1.35e-58 | 95 | 293 | 2 | 192 | AtlA is an autolysin found in Gram-positive lactic acid bacteria that degrades bacterial cell walls by catalyzing the hydrolysis of 1,4-beta-linkages between N-acetylmuramic acid and N-acetyl-D-glucosamine residues. This family includes the AtlA and Aml autolysins from Streptococcus mutans which have a C-terminal glycosyl hydrolase family 25 (GH25) catalytic domain as well as six tandem N-terminal repeats of the GBS (group B Streptococcus) Bsp-like peptidoglycan-binding domain. Other members of this family have one or more C-terminal peptidoglycan-binding domain(s) (SH3 or LysM) in addition to the GH25 domain. |
| NF033930 | pneumo_PspA | 2.65e-28 | 522 | 755 | 439 | 640 | pneumococcal surface protein A. The pneumococcal surface protein proteins, found in Streptococcus pneumoniae, are repetitive, with patterns of localized high sequence identity across pairs of proteins given different specific names that recombination may be presumed. This protein, PspA, has an N-terminal region that lacks a cross-wall-targeting YSIRK type extended signal peptide, in contrast to the closely related choline-binding protein CbpA which has a similar C-terminus but a YSIRK-containing region at the N-terminus. |
| NF033930 | pneumo_PspA | 7.13e-27 | 322 | 583 | 443 | 640 | pneumococcal surface protein A. The pneumococcal surface protein proteins, found in Streptococcus pneumoniae, are repetitive, with patterns of localized high sequence identity across pairs of proteins given different specific names that recombination may be presumed. This protein, PspA, has an N-terminal region that lacks a cross-wall-targeting YSIRK type extended signal peptide, in contrast to the closely related choline-binding protein CbpA which has a similar C-terminus but a YSIRK-containing region at the N-terminus. |
| COG5263 | COG5263 | 2.77e-25 | 551 | 772 | 87 | 309 | Glucan-binding domain (YG repeat) [Carbohydrate transport and metabolism]. |
| pfam01183 | Glyco_hydro_25 | 3.98e-25 | 97 | 279 | 1 | 178 | Glycosyl hydrolases family 25. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEN77492.1 | 5.15e-267 | 29 | 578 | 1 | 551 |
| AWZ39196.1 | 3.17e-76 | 74 | 321 | 199 | 444 |
| AWZ40161.1 | 3.17e-76 | 74 | 321 | 199 | 444 |
| QCQ03274.1 | 2.10e-75 | 80 | 312 | 205 | 433 |
| AUJ29432.1 | 9.51e-75 | 72 | 333 | 202 | 465 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
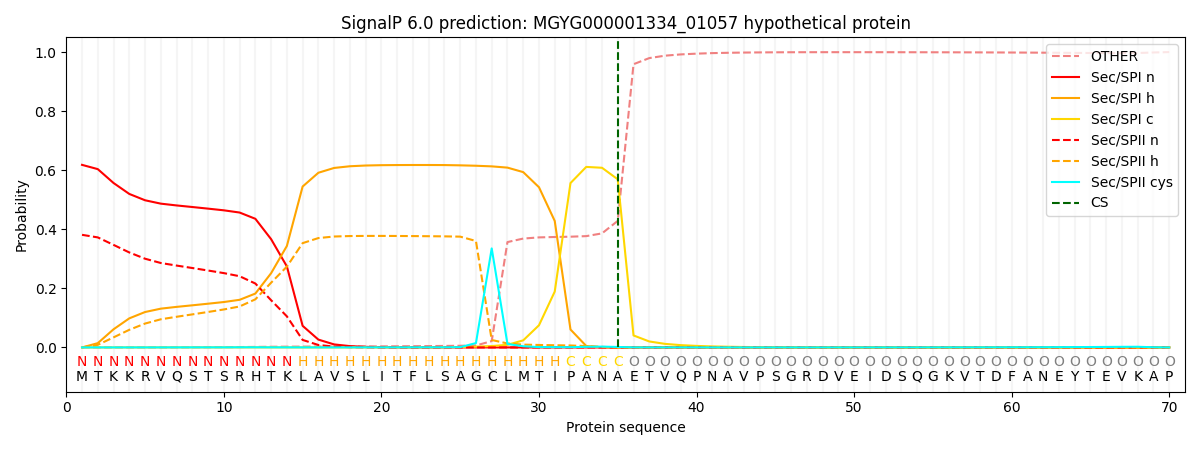
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000606 | 0.606725 | 0.391147 | 0.000858 | 0.000399 | 0.000247 |