You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001336_00718
You are here: Home > Sequence: MGYG000001336_00718
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
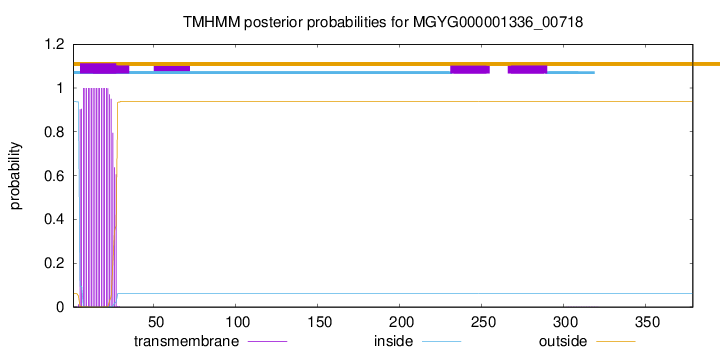
TMHMM annotations
Basic Information help
| Species | Limosilactobacillus reuteri_E | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Limosilactobacillus; Limosilactobacillus reuteri_E | |||||||||||
| CAZyme ID | MGYG000001336_00718 | |||||||||||
| CAZy Family | GH8 | |||||||||||
| CAZyme Description | Endoglucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 690297; End: 691436 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01270 | Glyco_hydro_8 | 6.05e-18 | 69 | 375 | 34 | 317 | Glycosyl hydrolases family 8. |
| PRK11097 | PRK11097 | 9.17e-14 | 52 | 370 | 32 | 345 | cellulase. |
| COG3405 | BcsZ | 1.09e-11 | 56 | 377 | 44 | 344 | Endo-1,4-beta-D-glucanase Y [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEI56910.1 | 2.25e-289 | 1 | 379 | 1 | 379 |
| AAY86852.1 | 2.25e-289 | 1 | 379 | 1 | 379 |
| QIZ04939.1 | 5.30e-288 | 1 | 379 | 1 | 379 |
| AGN98608.1 | 1.77e-286 | 1 | 379 | 1 | 379 |
| AMY14459.1 | 1.77e-286 | 1 | 379 | 1 | 379 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6VC5_A | 4.69e-06 | 69 | 285 | 33 | 231 | 1.6Angstrom Resolution Crystal Structure of endoglucanase from Komagataeibacter sucrofermentans [Komagataeibacter sucrofermentans] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
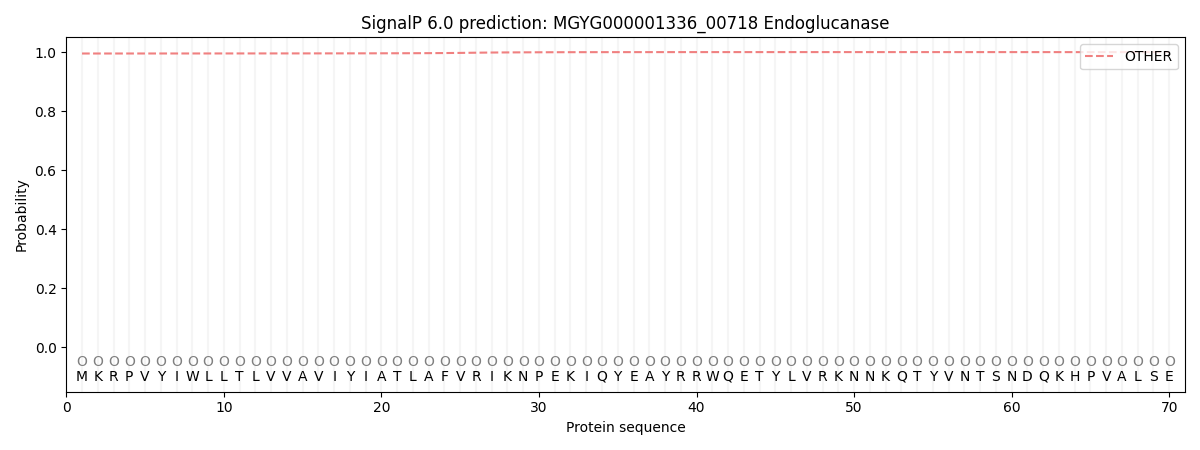
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.995513 | 0.004271 | 0.000106 | 0.000022 | 0.000012 | 0.000112 |