You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001337_01433
You are here: Home > Sequence: MGYG000001337_01433
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
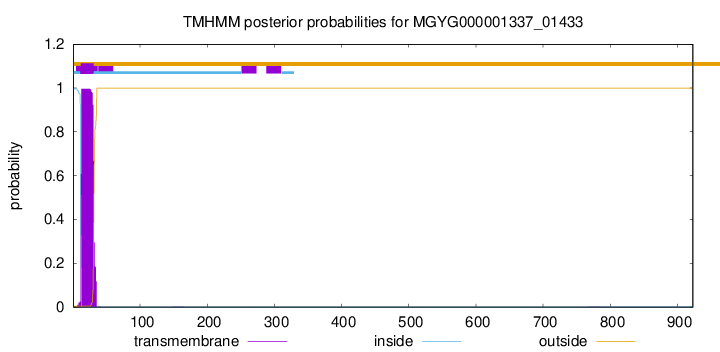
TMHMM annotations
Basic Information help
| Species | Bacteroides fragilis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides fragilis | |||||||||||
| CAZyme ID | MGYG000001337_01433 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1147138; End: 1149909 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 25 | 751 | 1.7e-78 | 0.773936170212766 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 2.84e-27 | 29 | 472 | 7 | 457 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 4.50e-27 | 28 | 328 | 6 | 296 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 1.30e-14 | 102 | 454 | 113 | 471 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam02837 | Glyco_hydro_2_N | 6.46e-11 | 37 | 201 | 4 | 168 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| pfam00703 | Glyco_hydro_2 | 8.95e-11 | 204 | 308 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANQ59584.1 | 0.0 | 1 | 923 | 1 | 923 |
| BAD47686.1 | 0.0 | 1 | 923 | 1 | 923 |
| QUU05975.1 | 0.0 | 1 | 923 | 1 | 923 |
| QRP91141.1 | 0.0 | 1 | 923 | 1 | 923 |
| CBW21485.1 | 0.0 | 1 | 923 | 1 | 923 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6SD0_A | 1.89e-23 | 32 | 471 | 37 | 478 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 6S6Z_A | 1.89e-23 | 32 | 471 | 36 | 477 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6D8G_A | 1.16e-20 | 88 | 431 | 74 | 426 | D341AD367A calcium binding mutant of Bacteroides uniformis beta-glucuronidase 2 [Bacteroides uniformis str. 3978 T3 ii],6D8G_B D341A D367A calcium binding mutant of Bacteroides uniformis beta-glucuronidase 2 [Bacteroides uniformis str. 3978 T3 ii] |
| 5UJ6_A | 2.62e-20 | 88 | 431 | 66 | 418 | CrystalStructure of Bacteroides Uniformis beta-glucuronidase [Bacteroides uniformis str. 3978 T3 ii],5UJ6_B Crystal Structure of Bacteroides Uniformis beta-glucuronidase [Bacteroides uniformis str. 3978 T3 ii],6NZG_A Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine [Bacteroides uniformis],6NZG_B Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine [Bacteroides uniformis] |
| 6D50_A | 2.64e-20 | 88 | 431 | 74 | 426 | Bacteroidesuniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone [Bacteroides uniformis str. 3978 T3 ii],6D50_B Bacteroides uniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone [Bacteroides uniformis str. 3978 T3 ii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56307 | 1.04e-22 | 32 | 471 | 37 | 478 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| P08236 | 3.92e-10 | 31 | 431 | 35 | 451 | Beta-glucuronidase OS=Homo sapiens OX=9606 GN=GUSB PE=1 SV=2 |
| O77695 | 1.54e-09 | 31 | 431 | 32 | 448 | Beta-glucuronidase (Fragment) OS=Chlorocebus aethiops OX=9534 GN=GUSB PE=2 SV=1 |
| Q8D4H3 | 1.94e-09 | 138 | 432 | 157 | 459 | Beta-galactosidase OS=Vibrio vulnificus (strain CMCP6) OX=216895 GN=lacZ PE=3 SV=2 |
| Q7MG04 | 5.75e-09 | 138 | 432 | 157 | 458 | Beta-galactosidase OS=Vibrio vulnificus (strain YJ016) OX=196600 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
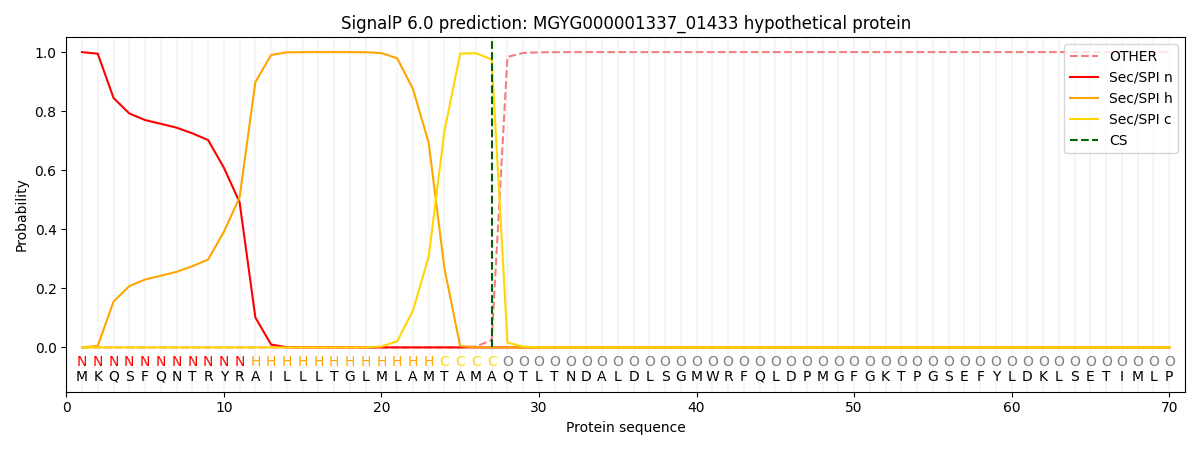
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000492 | 0.998513 | 0.000280 | 0.000241 | 0.000225 | 0.000194 |