You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001345_03615
You are here: Home > Sequence: MGYG000001345_03615
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides xylanisolvens | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides xylanisolvens | |||||||||||
| CAZyme ID | MGYG000001345_03615 | |||||||||||
| CAZy Family | CE6 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 76580; End: 77377 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE6 | 91 | 183 | 6.9e-27 | 0.98989898989899 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03629 | SASA | 3.47e-89 | 30 | 260 | 1 | 226 | Carbohydrate esterase, sialic acid-specific acetylesterase. The catalytic triad of this esterase enzyme comprises residues Ser127, His403 and Asp391 in UniProtKB:P70665. |
| cd01833 | XynB_like | 0.004 | 152 | 216 | 44 | 109 | SGNH_hydrolase subfamily, similar to Ruminococcus flavefaciens XynB. Most likely a secreted hydrolase with xylanase activity. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT32559.1 | 5.49e-194 | 1 | 265 | 1 | 265 |
| QDH54197.1 | 2.61e-192 | 1 | 265 | 1 | 265 |
| QUT78436.1 | 4.33e-191 | 1 | 265 | 1 | 265 |
| QDM11351.1 | 4.33e-191 | 1 | 265 | 1 | 265 |
| QRM98824.1 | 4.33e-191 | 1 | 265 | 1 | 265 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2APJ_A | 3.23e-19 | 21 | 259 | 9 | 256 | X-RayStructure of Protein from Arabidopsis Thaliana AT4G34215 at 1.6 Angstrom Resolution [Arabidopsis thaliana],2APJ_B X-Ray Structure of Protein from Arabidopsis Thaliana AT4G34215 at 1.6 Angstrom Resolution [Arabidopsis thaliana],2APJ_C X-Ray Structure of Protein from Arabidopsis Thaliana AT4G34215 at 1.6 Angstrom Resolution [Arabidopsis thaliana],2APJ_D X-Ray Structure of Protein from Arabidopsis Thaliana AT4G34215 at 1.6 Angstrom Resolution [Arabidopsis thaliana] |
| 1ZMB_A | 4.50e-17 | 34 | 254 | 5 | 220 | CrystalStructure of the Putative Acetylxylan Esterase from Clostridium acetobutylicum, Northeast Structural Genomics Target CaR6 [Clostridium acetobutylicum ATCC 824],1ZMB_B Crystal Structure of the Putative Acetylxylan Esterase from Clostridium acetobutylicum, Northeast Structural Genomics Target CaR6 [Clostridium acetobutylicum ATCC 824],1ZMB_C Crystal Structure of the Putative Acetylxylan Esterase from Clostridium acetobutylicum, Northeast Structural Genomics Target CaR6 [Clostridium acetobutylicum ATCC 824],1ZMB_D Crystal Structure of the Putative Acetylxylan Esterase from Clostridium acetobutylicum, Northeast Structural Genomics Target CaR6 [Clostridium acetobutylicum ATCC 824],1ZMB_E Crystal Structure of the Putative Acetylxylan Esterase from Clostridium acetobutylicum, Northeast Structural Genomics Target CaR6 [Clostridium acetobutylicum ATCC 824],1ZMB_F Crystal Structure of the Putative Acetylxylan Esterase from Clostridium acetobutylicum, Northeast Structural Genomics Target CaR6 [Clostridium acetobutylicum ATCC 824] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8L9J9 | 3.47e-19 | 21 | 259 | 9 | 256 | Probable carbohydrate esterase At4g34215 OS=Arabidopsis thaliana OX=3702 GN=At4g34215 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
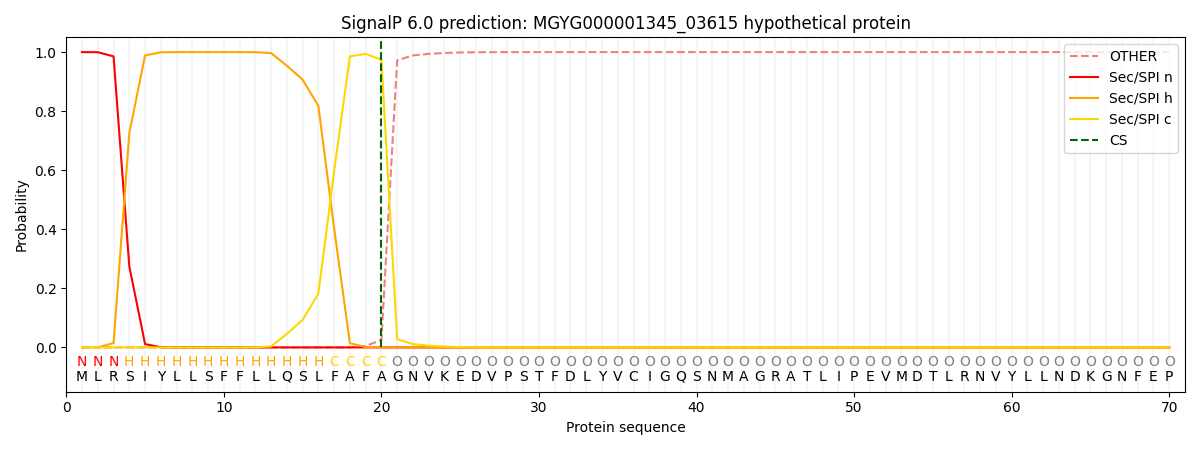
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000290 | 0.999063 | 0.000186 | 0.000146 | 0.000150 | 0.000147 |
