You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001345_03803
You are here: Home > Sequence: MGYG000001345_03803
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
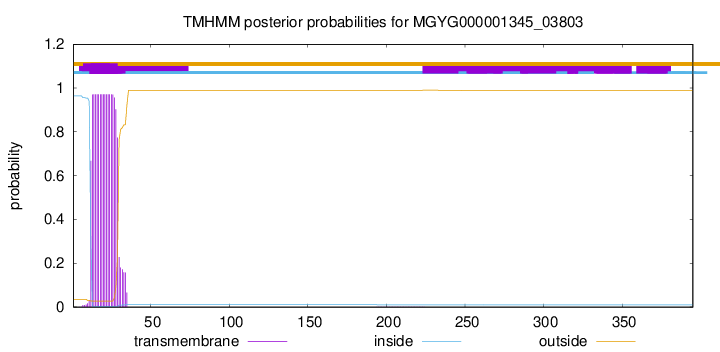
TMHMM annotations
Basic Information help
| Species | Bacteroides xylanisolvens | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides xylanisolvens | |||||||||||
| CAZyme ID | MGYG000001345_03803 | |||||||||||
| CAZy Family | CE15 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 357734; End: 358921 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE15 | 63 | 388 | 2.6e-19 | 0.9814126394052045 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0412 | DLH | 2.36e-07 | 125 | 260 | 7 | 140 | Dienelactone hydrolase [Secondary metabolites biosynthesis, transport and catabolism]. |
| COG1506 | DAP2 | 6.78e-05 | 130 | 388 | 376 | 615 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QMW85982.1 | 6.26e-200 | 9 | 387 | 2 | 383 |
| QQA08270.1 | 6.26e-200 | 9 | 387 | 2 | 383 |
| ALJ44193.1 | 6.26e-200 | 9 | 387 | 2 | 383 |
| QDH54042.1 | 6.26e-200 | 18 | 394 | 12 | 392 |
| AAO78713.1 | 6.26e-200 | 9 | 387 | 2 | 383 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7NN3_A | 2.85e-09 | 54 | 395 | 17 | 374 | ChainA, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B],7NN3_B Chain B, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B],7NN3_C Chain C, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B],7NN3_D Chain D, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
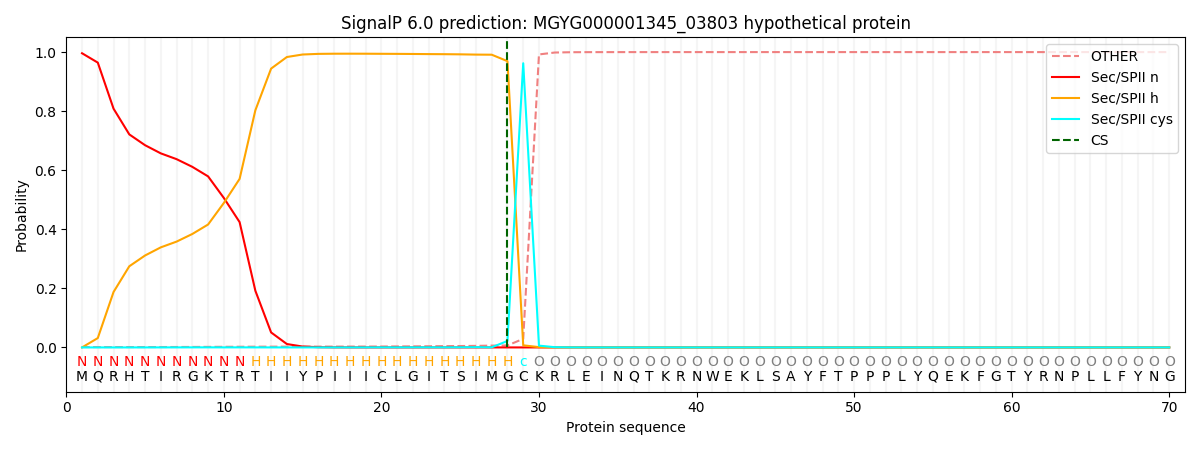
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001263 | 0.002236 | 0.996497 | 0.000003 | 0.000012 | 0.000003 |