You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001345_03807
You are here: Home > Sequence: MGYG000001345_03807
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides xylanisolvens | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides xylanisolvens | |||||||||||
| CAZyme ID | MGYG000001345_03807 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 365645; End: 367213 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 317 | 443 | 4.9e-25 | 0.3795379537953795 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4124 | ManB2 | 7.63e-12 | 312 | 476 | 169 | 332 | Beta-mannanase [Carbohydrate transport and metabolism]. |
| pfam02156 | Glyco_hydro_26 | 2.24e-07 | 215 | 409 | 50 | 235 | Glycosyl hydrolase family 26. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBK67924.1 | 0.0 | 1 | 522 | 1 | 522 |
| QRM99037.1 | 0.0 | 1 | 522 | 1 | 522 |
| QUT26858.1 | 0.0 | 1 | 522 | 1 | 522 |
| QGT73042.1 | 0.0 | 1 | 522 | 1 | 522 |
| QNL40569.1 | 0.0 | 1 | 522 | 1 | 522 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6HF2_A | 4.19e-08 | 324 | 456 | 193 | 315 | Thestructure of BoMan26B, a GH26 beta-mannanase from Bacteroides ovatus [Bacteroides ovatus ATCC 8483],6HF4_A The structure of BoMan26B, a GH26 beta-mannanase from Bacteroides ovatus, complexed with G1M4 [Bacteroides ovatus ATCC 8483] |
| 6Q75_A | 1.48e-06 | 324 | 404 | 169 | 252 | Thestructure of GH26A from Muricauda sp. MAR_2010_75 [Muricauda sp. MAR_2010_75],6Q75_B The structure of GH26A from Muricauda sp. MAR_2010_75 [Muricauda sp. MAR_2010_75] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
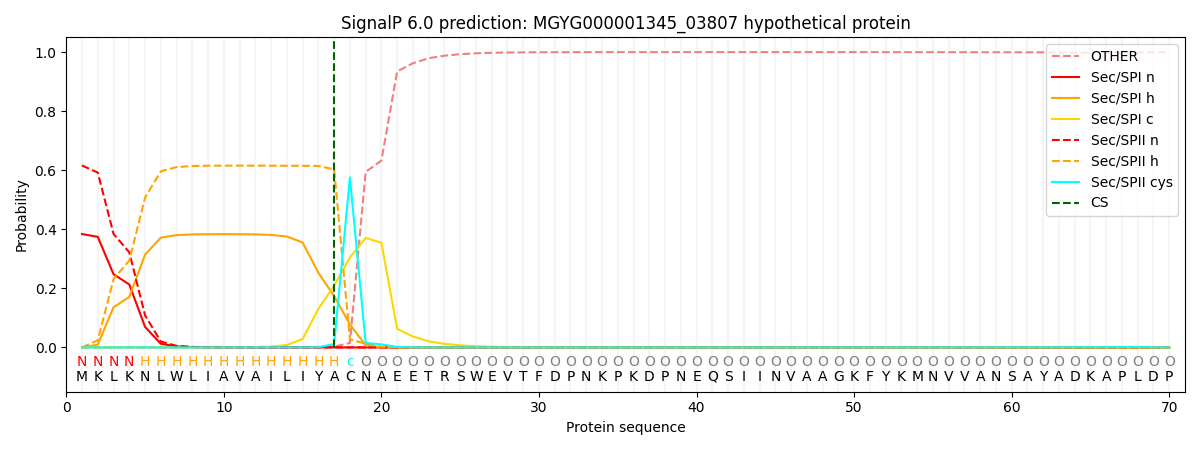
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000325 | 0.377601 | 0.621557 | 0.000181 | 0.000180 | 0.000153 |
