You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001346_02288
You are here: Home > Sequence: MGYG000001346_02288
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides uniformis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides uniformis | |||||||||||
| CAZyme ID | MGYG000001346_02288 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Xyloglucan-specific endo-beta-1,4-glucanase BoGH5A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1067986; End: 1069419 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 163 | 446 | 5.7e-83 | 0.9891304347826086 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 2.00e-44 | 148 | 411 | 1 | 236 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 4.32e-19 | 133 | 411 | 46 | 321 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| cd14948 | BACON | 1.79e-16 | 30 | 112 | 2 | 83 | Bacteroidetes-Associated Carbohydrate-binding (putative) Often N-terminal (BACON) domain. The BACON domain is found in diverse domain architectures and accociated with a wide variety of domains, including carbohydrate-active enzymes and proteases. It was named for its suggested function of carbohydrate binding; the latter was inferred from domain architectures, sequence conservation, and phyletic distribution. However, recent experimental data suggest that its primary function in Bacteroides ovatus endo-xyloglucanase BoGH5A is to distance the catalytic module from the cell surface and confer additional mobility to the catalytic domain for attack of the polysaccharide. No evidence for a direct role in carbohydrate binding could be found in that case. The large majority of BACON domains are found in Bacteroidetes. |
| pfam13004 | BACON | 8.94e-08 | 54 | 112 | 1 | 61 | Putative binding domain, N-terminal. The BACON (Bacteroidetes-Associated Carbohydrate-binding Often N-terminal) domain is an all-beta domain found in diverse architectures, principally in combination with carbohydrate-active enzymes and proteases. These architectures suggest a carbohydrate-binding function which is also supported by the nature of BACON's few conserved amino-acids. The phyletic distribution of BACON and other data tentatively suggest that it may frequently function to bind mucin. Further work with the characterized structure of a member of glycoside hydrolase family 5 enzyme, Structure 3ZMR, has found no evidence for carbohydrate-binding for this domain. |
| pfam19190 | BACON_2 | 6.92e-05 | 30 | 114 | 2 | 91 | Viral BACON domain. This family represents a distinct class of BACON domains found in crAss-like phages, the most common viral family in the human gut, in which they are found in tail fiber genes. This suggests they may play a role in phage-host interactions. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT61941.1 | 0.0 | 1 | 477 | 1 | 477 |
| QUU00756.1 | 0.0 | 1 | 477 | 1 | 477 |
| QQA30413.1 | 0.0 | 1 | 477 | 1 | 477 |
| BBK86024.1 | 0.0 | 1 | 477 | 1 | 477 |
| QUT65254.1 | 0.0 | 1 | 477 | 4 | 480 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6XRK_A | 7.01e-68 | 130 | 470 | 23 | 370 | GH5-4broad specificity endoglucanase from an uncultured bovine rumen ciliate [uncultured bovine rumen ciliate],6XRK_B GH5-4 broad specificity endoglucanase from an uncultured bovine rumen ciliate [uncultured bovine rumen ciliate] |
| 1EDG_A | 1.29e-55 | 135 | 468 | 26 | 372 | SingleCrystal Structure Determination Of The Catalytic Domain Of Celcca Carried Out At 15 Degree C [Ruminiclostridium cellulolyticum H10] |
| 4NF7_A | 9.27e-54 | 133 | 468 | 10 | 361 | Crystalstructure of the GH5 family catalytic domain of Endo-1,4-beta-glucanase Cel5C from Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316] |
| 4IM4_A | 1.26e-53 | 128 | 452 | 4 | 305 | ChainA, Endoglucanase E [Acetivibrio thermocellus],4IM4_B Chain B, Endoglucanase E [Acetivibrio thermocellus],4IM4_C Chain C, Endoglucanase E [Acetivibrio thermocellus],4IM4_D Chain D, Endoglucanase E [Acetivibrio thermocellus],4IM4_E Chain E, Endoglucanase E [Acetivibrio thermocellus],4IM4_F Chain F, Endoglucanase E [Acetivibrio thermocellus] |
| 4X0V_A | 2.10e-53 | 133 | 476 | 42 | 396 | Structureof a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_B Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_C Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_D Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_E Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_F Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_G Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_H Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P17901 | 6.45e-54 | 135 | 468 | 51 | 397 | Endoglucanase A OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCA PE=1 SV=1 |
| P20847 | 5.12e-51 | 133 | 468 | 41 | 392 | Endoglucanase 1 OS=Butyrivibrio fibrisolvens OX=831 GN=end1 PE=3 SV=1 |
| P28623 | 5.53e-51 | 123 | 461 | 35 | 362 | Endoglucanase D OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engD PE=1 SV=2 |
| A7LXT7 | 1.62e-50 | 4 | 445 | 20 | 473 | Xyloglucan-specific endo-beta-1,4-glucanase BoGH5A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02653 PE=1 SV=1 |
| P23660 | 5.91e-50 | 128 | 456 | 24 | 348 | Endoglucanase A OS=Ruminococcus albus OX=1264 GN=celA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
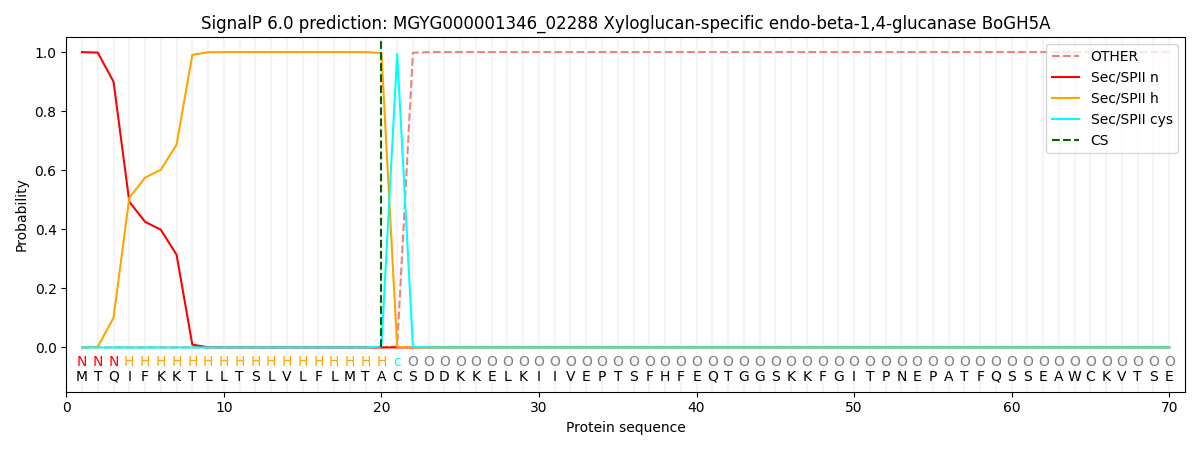
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000056 | 0.000000 | 0.000000 | 0.000000 |
