You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001350_01959
Basic Information
help
| Species |
Fusobacterium periodonticum_B
|
| Lineage |
Bacteria; Fusobacteriota; Fusobacteriia; Fusobacteriales; Fusobacteriaceae; Fusobacterium; Fusobacterium periodonticum_B
|
| CAZyme ID |
MGYG000001350_01959
|
| CAZy Family |
GT83 |
| CAZyme Description |
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 519 |
|
61723.57 |
9.8269 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001350 |
2454675 |
Isolate |
not provided |
not provided |
|
| Gene Location |
Start: 836240;
End: 837799
Strand: +
|
No EC number prediction in MGYG000001350_01959.
| Family |
Start |
End |
Evalue |
family coverage |
| GT83 |
4 |
431 |
3.4e-52 |
0.8111111111111111 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG1807
|
ArnT |
1.37e-57 |
1 |
519 |
2 |
529 |
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231
|
PMT_2 |
1.36e-10 |
63 |
223 |
3 |
160 |
Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| PRK13279
|
arnT |
2.16e-07 |
4 |
224 |
3 |
224 |
lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
| COG1928
|
PMT1 |
0.005 |
47 |
230 |
56 |
269 |
Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
This protein is predicted as OTHER
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.994830
|
0.004817
|
0.000072
|
0.000036
|
0.000027
|
0.000241
|
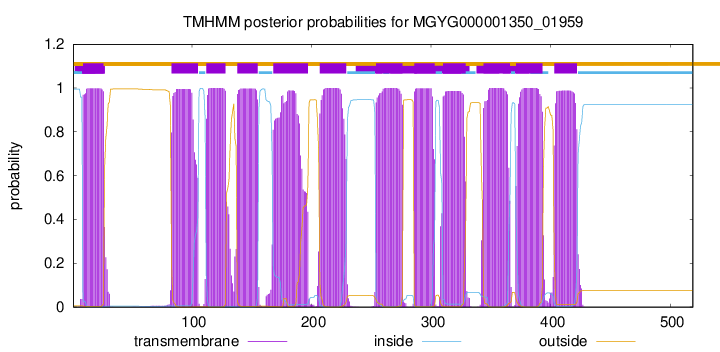
| start |
end |
| 9 |
26 |
| 83 |
105 |
| 112 |
128 |
| 138 |
155 |
| 168 |
197 |
| 207 |
229 |
| 254 |
276 |
| 286 |
303 |
| 310 |
329 |
| 344 |
366 |
| 371 |
393 |
| 403 |
422 |


