You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001356_00228
You are here: Home > Sequence: MGYG000001356_00228
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
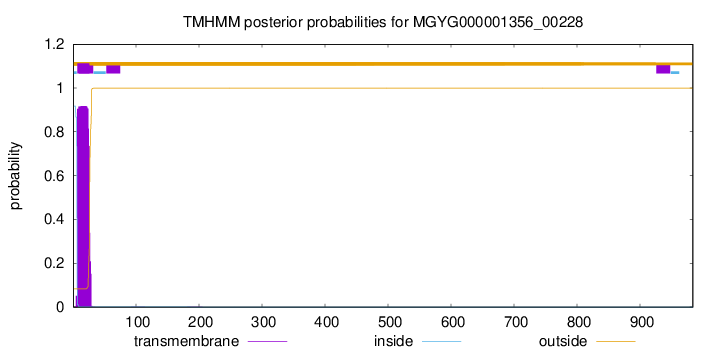
TMHMM annotations
Basic Information help
| Species | Lawsonibacter sp000177015 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; Lawsonibacter; Lawsonibacter sp000177015 | |||||||||||
| CAZyme ID | MGYG000001356_00228 | |||||||||||
| CAZy Family | SLH | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 247092; End: 250046 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM54 | 192 | 302 | 1.5e-32 | 0.9649122807017544 |
| SLH | 85 | 126 | 1.3e-16 | 0.9761904761904762 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02368 | Big_2 | 5.43e-15 | 420 | 495 | 2 | 77 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| COG5492 | YjdB | 5.32e-14 | 408 | 500 | 171 | 264 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| pfam00395 | SLH | 8.89e-11 | 85 | 126 | 1 | 42 | S-layer homology domain. |
| smart00635 | BID_2 | 4.15e-09 | 420 | 494 | 4 | 81 | Bacterial Ig-like domain 2. |
| pfam00395 | SLH | 6.63e-08 | 146 | 186 | 1 | 41 | S-layer homology domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOX65359.1 | 4.08e-130 | 24 | 937 | 26 | 905 |
| AEV69543.1 | 9.44e-119 | 1 | 940 | 1 | 952 |
| QKS47579.1 | 3.98e-109 | 1 | 755 | 5 | 844 |
| ASA25822.1 | 1.60e-68 | 26 | 497 | 48 | 528 |
| AWP25495.1 | 7.48e-57 | 28 | 330 | 44 | 361 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3PYW_A | 1.23e-11 | 33 | 218 | 12 | 201 | Thestructure of the SLH domain from B. anthracis surface array protein at 1.8A [Bacillus anthracis] |
| 6BT4_A | 2.22e-11 | 33 | 188 | 33 | 189 | Crystalstructure of the SLH domain of Sap from Bacillus anthracis in complex with a pyruvylated SCWP unit [Bacillus anthracis] |
| 5GZT_A | 5.42e-08 | 202 | 251 | 49 | 98 | CrystalStructure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery [Paenibacillus sp. FPU-7] |
| 4AQ1_A | 2.23e-06 | 84 | 193 | 3 | 108 | Structureof the SbsB S-layer protein of Geobacillus stearothermophilus PV72p2 in complex with nanobody KB6 [Geobacillus stearothermophilus] |
| 4AQ1_C | 2.23e-06 | 84 | 193 | 3 | 108 | Structureof the SbsB S-layer protein of Geobacillus stearothermophilus PV72p2 in complex with nanobody KB6 [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P38537 | 5.36e-16 | 32 | 202 | 40 | 213 | Surface-layer 125 kDa protein OS=Lysinibacillus sphaericus OX=1421 PE=3 SV=1 |
| C6CRV0 | 1.52e-14 | 34 | 195 | 1291 | 1459 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
| Q06853 | 1.06e-12 | 45 | 186 | 481 | 629 | Cell surface glycoprotein 2 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=Cthe_3079 PE=1 SV=1 |
| P49051 | 2.08e-12 | 4 | 335 | 7 | 331 | S-layer protein sap OS=Bacillus anthracis OX=1392 GN=sap PE=1 SV=1 |
| Q06852 | 2.22e-12 | 48 | 186 | 2104 | 2255 | Cell surface glycoprotein 1 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=olpB PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
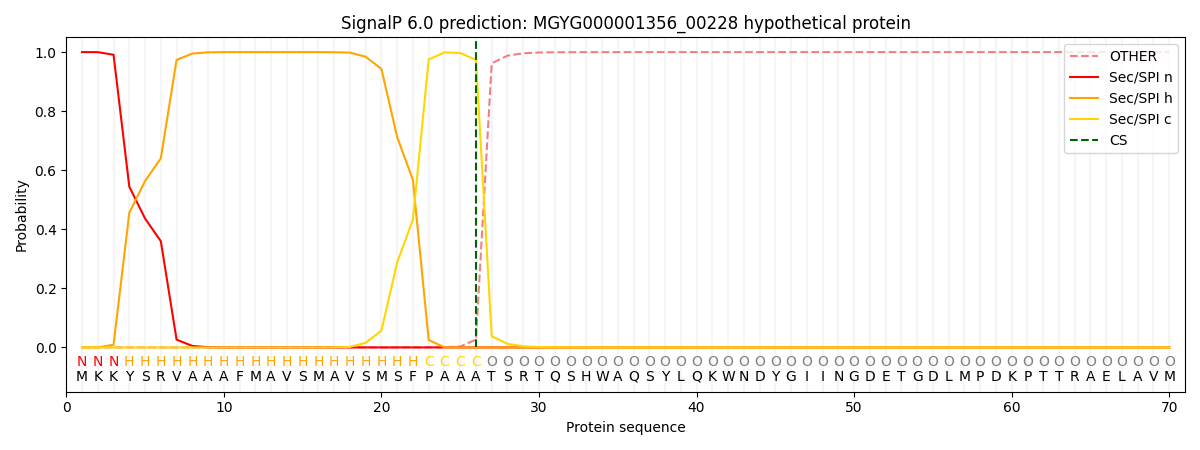
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000354 | 0.998825 | 0.000216 | 0.000222 | 0.000200 | 0.000164 |