You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001359_01529
You are here: Home > Sequence: MGYG000001359_01529
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bilophila wadsworthia | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Desulfobacterota; Desulfovibrionia; Desulfovibrionales; Desulfovibrionaceae; Bilophila; Bilophila wadsworthia | |||||||||||
| CAZyme ID | MGYG000001359_01529 | |||||||||||
| CAZy Family | GH103 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1670415; End: 1671422 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH103 | 122 | 305 | 4.6e-44 | 0.5254237288135594 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13406 | SLT_2 | 1.36e-49 | 122 | 328 | 54 | 232 | Transglycosylase SLT domain. This family is related to the SLT domain pfam01464. |
| COG2951 | MltB | 1.15e-34 | 124 | 331 | 95 | 281 | Membrane-bound lytic murein transglycosylase B [Cell wall/membrane/envelope biogenesis]. |
| PRK10760 | PRK10760 | 1.39e-23 | 126 | 304 | 120 | 270 | murein hydrolase B; Provisional |
| cd13399 | Slt35-like | 3.03e-23 | 245 | 329 | 22 | 108 | Slt35-like lytic transglycosylase. Lytic transglycosylase similar to Escherichia coli lytic transglycosylase Slt35 and Pseudomonas aeruginosa Sltb1. Lytic transglycosylase (LT) catalyzes the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc) as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. Proteins similar to this this family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, as well as the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADP85790.1 | 1.03e-99 | 33 | 334 | 45 | 334 |
| AAS95158.1 | 1.03e-99 | 33 | 334 | 45 | 334 |
| ABM29301.1 | 4.15e-99 | 33 | 334 | 45 | 334 |
| AMD89133.1 | 1.33e-90 | 47 | 328 | 111 | 383 |
| ATD81641.1 | 1.05e-89 | 30 | 328 | 84 | 375 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1LTM_A | 1.71e-18 | 126 | 299 | 81 | 226 | AcceleratedX-ray Structure Elucidation Of A 36 Kda Muramidase/transglycosylase Using Warp [Escherichia coli] |
| 1D0K_A | 1.75e-18 | 126 | 299 | 83 | 228 | ChainA, 35KD SOLUBLE LYTIC TRANSGLYCOSYLASE [Escherichia coli],1D0L_A Chain A, 35KD SOLUBLE LYTIC TRANSGLYCOSYLASE [Escherichia coli],1D0M_A Chain A, 35KD SOLUBLE LYTIC TRANSGLYCOSYLASE [Escherichia coli],1QDR_A Chain A, LYTIC MUREIN TRANSGLYCOSYLASE B [Escherichia coli],1QDT_A Chain A, LYTIC MUREIN TRANSGLYCOSYLASE B [Escherichia coli],1QUS_A Chain A, LYTIC MUREIN TRANSGLYCOSYLASE B [Escherichia coli],1QUT_A Chain A, LYTIC MUREIN TRANSGLYCOSYLASE B [Escherichia coli] |
| 5O8X_A | 3.88e-16 | 141 | 297 | 74 | 205 | TheX-ray Structure of Catenated Lytic Transglycosylase SltB1 from Pseudomonas aeruginosa [Pseudomonas aeruginosa],5O8X_B The X-ray Structure of Catenated Lytic Transglycosylase SltB1 from Pseudomonas aeruginosa [Pseudomonas aeruginosa] |
| 4ANR_A | 4.62e-16 | 141 | 297 | 91 | 222 | Crystalstructure of soluble lytic Transglycosylase SltB1 from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1] |
| 5ANZ_A | 1.51e-09 | 124 | 297 | 102 | 247 | CrystalStructure of SltB3 from Pseudomonas aeruginosa. [Pseudomonas aeruginosa],5AO7_A Crystal Structure of SltB3 from Pseudomonas aeruginosa in complex with NAG-anhNAM-pentapeptide [Pseudomonas aeruginosa] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P41052 | 1.42e-17 | 126 | 299 | 122 | 267 | Membrane-bound lytic murein transglycosylase B OS=Escherichia coli (strain K12) OX=83333 GN=mltB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
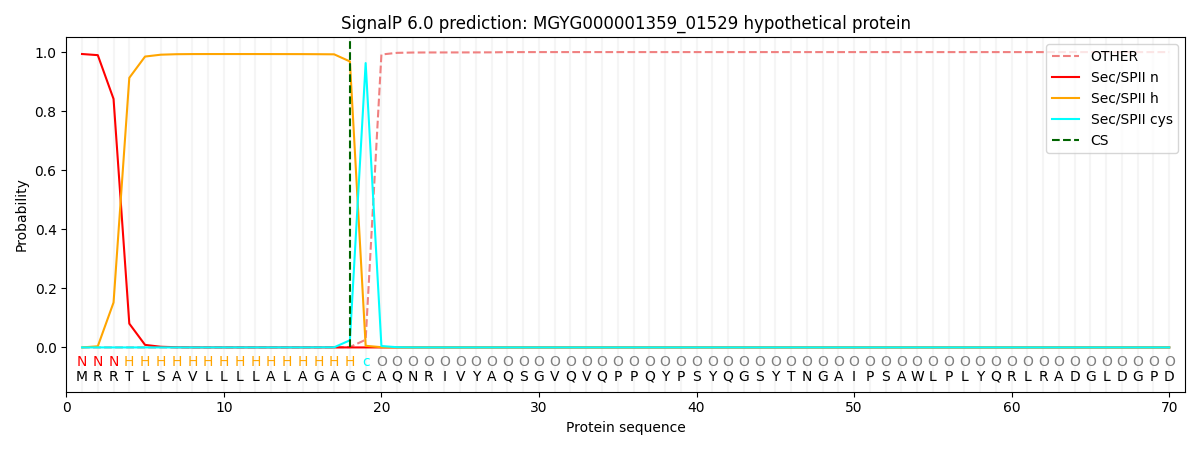
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000009 | 0.006325 | 0.993706 | 0.000003 | 0.000004 | 0.000003 |
