You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001364_00573
You are here: Home > Sequence: MGYG000001364_00573
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
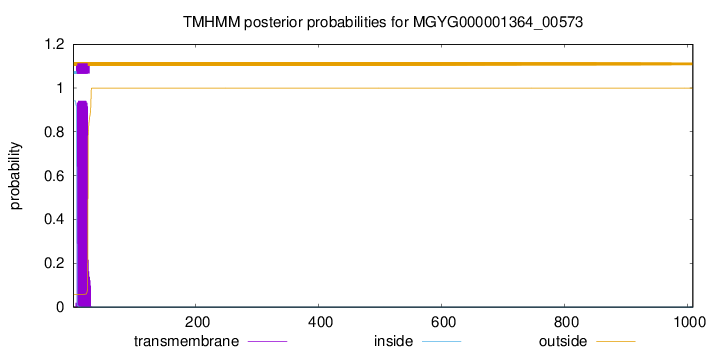
TMHMM annotations
Basic Information help
| Species | Phocaeicola plebeius | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola plebeius | |||||||||||
| CAZyme ID | MGYG000001364_00573 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 491917; End: 494946 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 245 | 1005 | 2.6e-197 | 0.8847087378640777 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 0.0 | 42 | 788 | 2 | 869 | alpha-L-rhamnosidase. |
| PRK10150 | PRK10150 | 6.65e-04 | 833 | 941 | 12 | 133 | beta-D-glucuronidase; Provisional |
| pfam02837 | Glyco_hydro_2_N | 0.001 | 854 | 951 | 47 | 144 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QEM10488.1 | 0.0 | 22 | 1004 | 10 | 998 |
| AWO01984.1 | 0.0 | 30 | 1008 | 14 | 990 |
| QEC76244.1 | 0.0 | 36 | 1007 | 26 | 999 |
| BAU55722.1 | 0.0 | 27 | 1004 | 22 | 1002 |
| ASU36259.1 | 0.0 | 36 | 1004 | 18 | 990 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q2F_A | 4.38e-130 | 30 | 1007 | 36 | 1141 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
| 5MQM_A | 6.13e-113 | 16 | 1002 | 9 | 1096 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A | 1.63e-112 | 16 | 1002 | 9 | 1096 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 6D89_A | 1.66e-07 | 861 | 942 | 86 | 168 | Bacteroidesuniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis],6D89_B Bacteroides uniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis],6D89_C Bacteroides uniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis],6D89_D Bacteroides uniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis] |
| 6D1N_A | 1.67e-07 | 861 | 942 | 94 | 176 | Apostructure of Bacteroides uniformis Beta-glucuronidase 1 [Bacteroides uniformis],6D1N_B Apo structure of Bacteroides uniformis Beta-glucuronidase 1 [Bacteroides uniformis],6D41_A Bacteriodes uniformis beta-glucuronidase 1 bound to D-glucaro-1,5-lactone [Bacteroides uniformis],6D41_B Bacteriodes uniformis beta-glucuronidase 1 bound to D-glucaro-1,5-lactone [Bacteroides uniformis],6D6W_A Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D6W_B Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D6W_C Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D6W_D Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D7F_A Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_B Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_C Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_D Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_E Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_F Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KNA8 | 1.18e-171 | 22 | 1004 | 14 | 1155 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
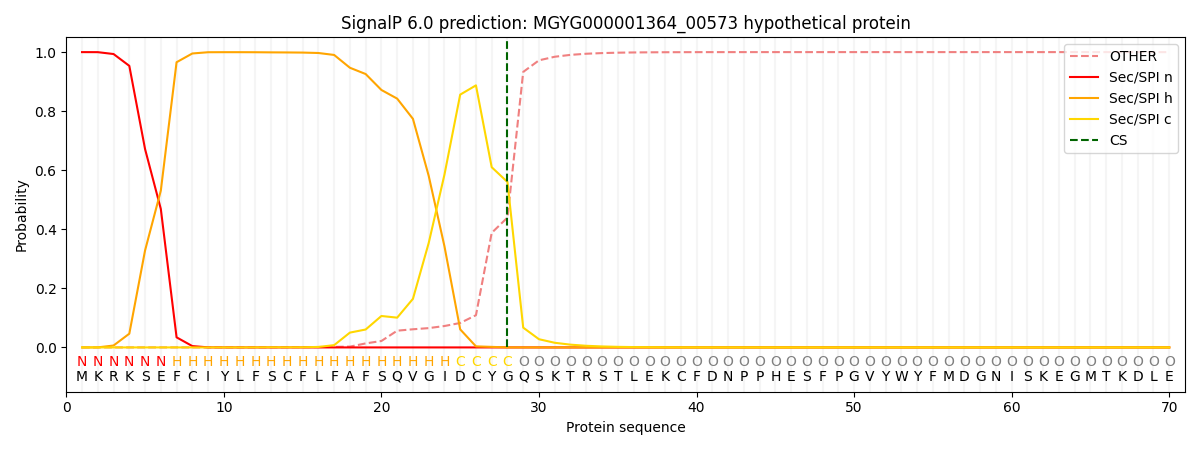
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000871 | 0.998348 | 0.000245 | 0.000187 | 0.000170 | 0.000165 |