You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001364_00579
You are here: Home > Sequence: MGYG000001364_00579
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
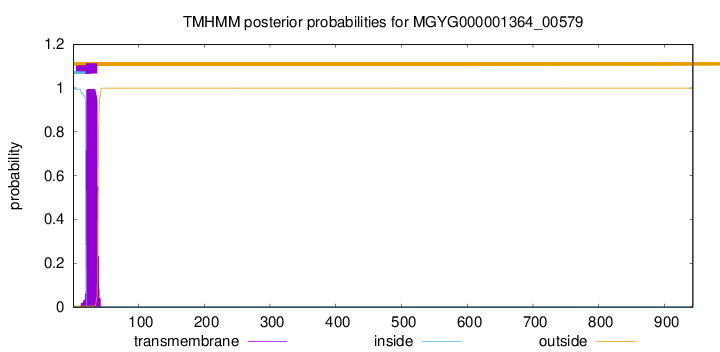
TMHMM annotations
Basic Information help
| Species | Phocaeicola plebeius | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola plebeius | |||||||||||
| CAZyme ID | MGYG000001364_00579 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 509788; End: 512619 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 38 | 501 | 8.3e-63 | 0.48936170212765956 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 2.78e-22 | 41 | 455 | 12 | 407 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 4.18e-16 | 38 | 348 | 9 | 296 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 5.15e-16 | 37 | 479 | 37 | 471 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam02837 | Glyco_hydro_2_N | 3.02e-08 | 44 | 190 | 4 | 135 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| pfam00703 | Glyco_hydro_2 | 9.70e-08 | 227 | 328 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRP90536.1 | 0.0 | 41 | 935 | 22 | 916 |
| QTO24394.1 | 0.0 | 28 | 943 | 6 | 924 |
| AUI46175.1 | 0.0 | 28 | 943 | 6 | 924 |
| AKA51553.1 | 0.0 | 41 | 935 | 22 | 916 |
| BAC56907.2 | 0.0 | 41 | 935 | 22 | 916 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7SF2_A | 3.24e-15 | 124 | 495 | 90 | 470 | ChainA, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_B Chain B, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_C Chain C, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_D Chain D, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_E Chain E, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_F Chain F, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838] |
| 6ECA_A | 4.57e-12 | 118 | 455 | 90 | 440 | Lactobacillusrhamnosus Beta-glucuronidase [Lacticaseibacillus rhamnosus],6ECA_B Lactobacillus rhamnosus Beta-glucuronidase [Lacticaseibacillus rhamnosus] |
| 6ED2_A | 2.40e-11 | 118 | 455 | 89 | 442 | ChainA, Glycosyl hydrolase family 2, TIM barrel domain protein [Faecalibacterium duncaniae] |
| 6D8G_A | 2.95e-11 | 117 | 508 | 81 | 501 | D341AD367A calcium binding mutant of Bacteroides uniformis beta-glucuronidase 2 [Bacteroides uniformis str. 3978 T3 ii],6D8G_B D341A D367A calcium binding mutant of Bacteroides uniformis beta-glucuronidase 2 [Bacteroides uniformis str. 3978 T3 ii] |
| 6LEM_B | 3.03e-11 | 118 | 455 | 61 | 412 | ChainB, Beta-D-glucuronidase [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q02603 | 1.59e-15 | 124 | 517 | 132 | 526 | Beta-galactosidase large subunit OS=Leuconostoc lactis OX=1246 GN=lacL PE=1 SV=1 |
| P06864 | 3.27e-13 | 124 | 544 | 113 | 546 | Evolved beta-galactosidase subunit alpha OS=Escherichia coli (strain K12) OX=83333 GN=ebgA PE=1 SV=4 |
| O07684 | 2.51e-11 | 37 | 479 | 38 | 490 | Beta-galactosidase large subunit OS=Lactobacillus acidophilus (strain ATCC 700396 / NCK56 / N2 / NCFM) OX=272621 GN=lacL PE=3 SV=2 |
| P05804 | 1.66e-10 | 118 | 455 | 63 | 414 | Beta-glucuronidase OS=Escherichia coli (strain K12) OX=83333 GN=uidA PE=1 SV=2 |
| Q59140 | 3.42e-09 | 124 | 454 | 118 | 434 | Beta-galactosidase OS=Arthrobacter sp. (strain B7) OX=86041 GN=lacZ PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
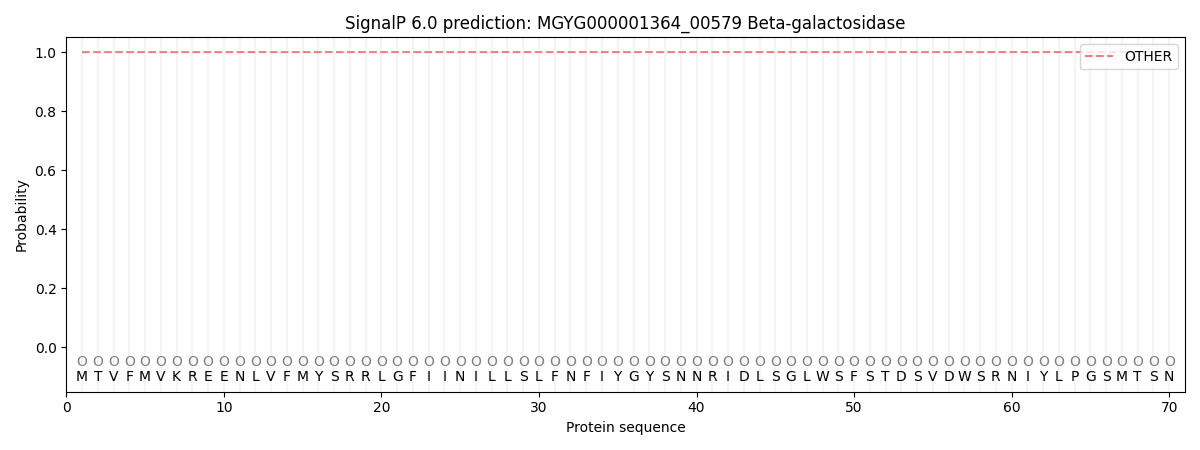
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999997 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |