You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001364_01559
You are here: Home > Sequence: MGYG000001364_01559
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola plebeius | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola plebeius | |||||||||||
| CAZyme ID | MGYG000001364_01559 | |||||||||||
| CAZy Family | GH86 | |||||||||||
| CAZyme Description | Beta-porphyranase A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 105011; End: 106834 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH86 | 28 | 599 | 6.9e-167 | 0.9898477157360406 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd21510 | agarase_cat | 4.59e-102 | 50 | 379 | 1 | 321 | alpha-beta barrel catalytic domain of agarase, such as GH86-like endo-acting agarases identified in non-marine organisms. Typically, agarases (E.C. 3.2.1.81) are found in ocean-dwelling bacteria since agarose is a principle component of red algae cell wall polysaccharides. Agarose is a linear polymer of alternating D-galactose and 3,6-anhydro-L-galactopyranose. Endo-acting agarases, such as glycoside hydrolase 16 (GH16) and GH86 hydrolyze internal beta-1,4 linkages. GH86-like endo-acting agarase of this protein family has been identified in the human intestinal bacterium Bacteroides uniformis. This acquired metabolic pathway, as demonstrated by the prevalence of agar-specific genetic cluster called polysaccharide utilization loci (PULs), varies considerably between human populations, being much more prevalent in a Japanese sample than in North America, European, or Chinese samples. Agarase activity was also identified in the non-marine bacterium Cellvibrio sp. |
| pfam18040 | BPA_C | 6.44e-34 | 506 | 598 | 2 | 94 | beta porphyranase A C-terminal. This is the C-terminal domain found in Bacteroides plebeius of proteins such as beta-porphyranase A (BPA), a beta-galactanase that cleaves the beta-1,4 glycosidic bond. Porphyranase degrade red seaweed glycans. This domain adopts a beta sandwich shape. |
| pfam18206 | Porphyrn_cat_1 | 6.70e-22 | 394 | 496 | 1 | 105 | Porphyranase catalytic subdomain 1. This domain is found in porphyranase protein present in Bacteroides plebeius. Porphyranase breaks down porphyran during digestion of red seaweed glycans. It is worth noting that red seaweed glycans contain sulfate esters that are absent in terrestrial plants. This domain makes up part of the catalytic domain of the porphyranase protein. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| EDY95428.1 | 0.0 | 34 | 607 | 1 | 574 |
| EDY95427.1 | 9.68e-250 | 32 | 601 | 28 | 595 |
| ANQ51878.1 | 9.09e-183 | 28 | 605 | 29 | 608 |
| QWG04144.1 | 7.19e-182 | 28 | 605 | 29 | 608 |
| AUP78946.1 | 4.49e-180 | 28 | 605 | 22 | 601 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4AW7_A | 4.47e-250 | 33 | 601 | 23 | 588 | ChainA, GH86A BETA-PORPHYRANASE [Phocaeicola plebeius] |
| 5TA1_A | 2.29e-83 | 30 | 606 | 25 | 647 | Crystalstructure of BuGH86wt [Bacteroides uniformis],5TA5_A Crystal structure of BuGH86wt in complex with neoagarooctaose [Bacteroides uniformis],5TA5_B Crystal structure of BuGH86wt in complex with neoagarooctaose [Bacteroides uniformis] |
| 5TA0_A | 6.25e-83 | 30 | 606 | 25 | 647 | Crystalstructure of BuGH86E322Q in complex with neoagarooctaose [Bacteroides uniformis],5TA0_B Crystal structure of BuGH86E322Q in complex with neoagarooctaose [Bacteroides uniformis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B5CY96 | 1.94e-250 | 32 | 601 | 28 | 595 | Beta-porphyranase A OS=Phocaeicola plebeius (strain DSM 17135 / JCM 12973 / M2) OX=484018 GN=BACPLE_01693 PE=1 SV=1 |
| P13734 | 3.04e-32 | 153 | 420 | 192 | 456 | Beta-agarase OS=Pseudoalteromonas atlantica OX=288 GN=agrA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
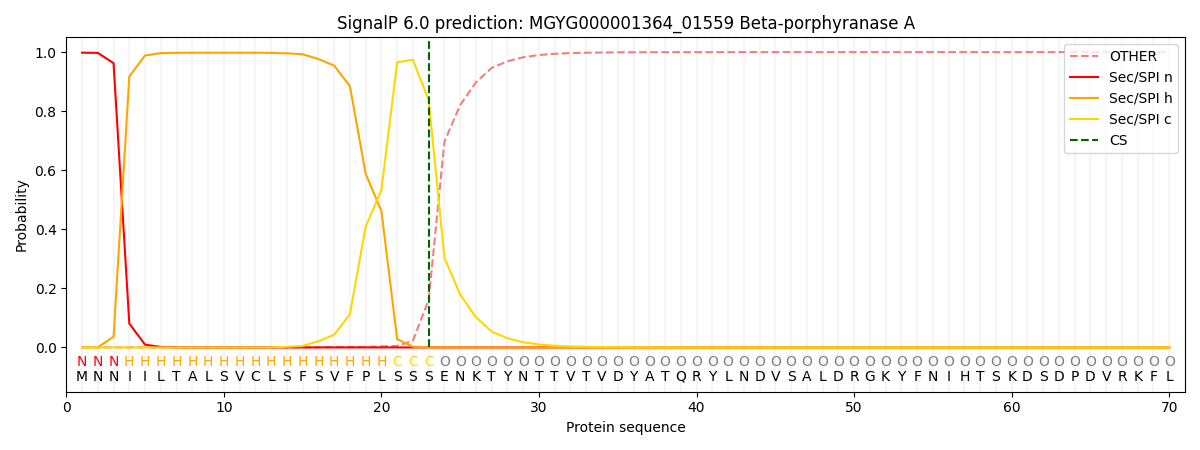
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000768 | 0.995872 | 0.002758 | 0.000197 | 0.000195 | 0.000188 |
