You are browsing environment: HUMAN GUT
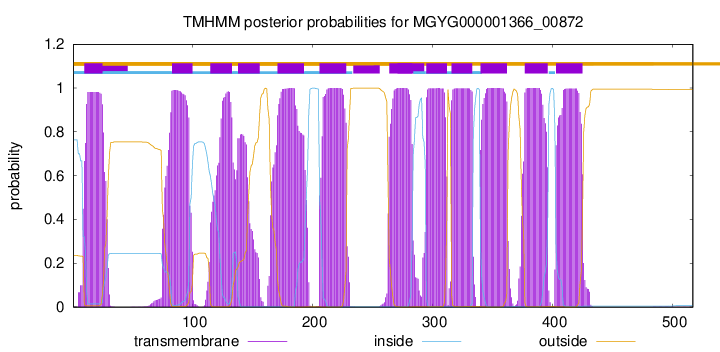
MGYG000001366_00872
Basic Information
help
Species
Succinatimonas hippei
Lineage
Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Succinivibrionaceae; Succinatimonas; Succinatimonas hippei
CAZyme ID
MGYG000001366_00872
CAZy Family
GT83
CAZyme Description
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000001366
2304998
Isolate
not provided
Asia
Gene Location
Start: 20311;
End: 21864
Strand: -
No EC number prediction in MGYG000001366_00872.
Family
Start
End
Evalue
family coverage
GT83
13
460
5.8e-69
0.8481481481481481
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
COG1807
ArnT
7.19e-22
13
352
15
364
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis].
more
PRK13279
arnT
2.45e-07
19
357
20
366
lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase.
more
pfam13231
PMT_2
0.003
61
209
1
144
Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
5EZM_A
6.93e-06
25
479
51
509
CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34]
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
O67270
7.48e-13
15
325
5
313
Uncharacterized protein aq_1220 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1220 PE=3 SV=1
more
C5BDQ8
8.77e-09
32
322
31
321
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Edwardsiella ictaluri (strain 93-146) OX=634503 GN=arnT PE=3 SV=1
more
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.999904
0.000107
0.000001
0.000000
0.000000
0.000001
start
end
10
25
83
100
115
133
138
156
171
193
206
228
264
283
295
312
316
333
340
362
377
396
403
425