You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001368_00166
You are here: Home > Sequence: MGYG000001368_00166
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Muricomes fissicatena_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Muricomes; Muricomes fissicatena_A | |||||||||||
| CAZyme ID | MGYG000001368_00166 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 201652; End: 206712 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH95 | 607 | 1286 | 7.2e-53 | 0.8642659279778393 |
| CBM32 | 494 | 625 | 1.7e-16 | 0.8790322580645161 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 5.30e-11 | 499 | 625 | 13 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 3.15e-09 | 146 | 286 | 1 | 124 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 0.001 | 503 | 628 | 28 | 142 | Substituted updates: Jan 31, 2002 |
| pfam17132 | Glyco_hydro_106 | 0.002 | 307 | 461 | 169 | 318 | alpha-L-rhamnosidase. |
| PRK02224 | PRK02224 | 0.003 | 1334 | 1479 | 428 | 558 | DNA double-strand break repair Rad50 ATPase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHW32096.1 | 1.02e-205 | 40 | 1333 | 42 | 1084 |
| AWS40658.1 | 3.00e-150 | 530 | 1330 | 173 | 926 |
| QXE33659.1 | 3.43e-142 | 527 | 1330 | 197 | 952 |
| BCB76148.1 | 1.26e-123 | 474 | 1330 | 81 | 879 |
| QKW17790.1 | 5.86e-85 | 636 | 1329 | 133 | 762 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
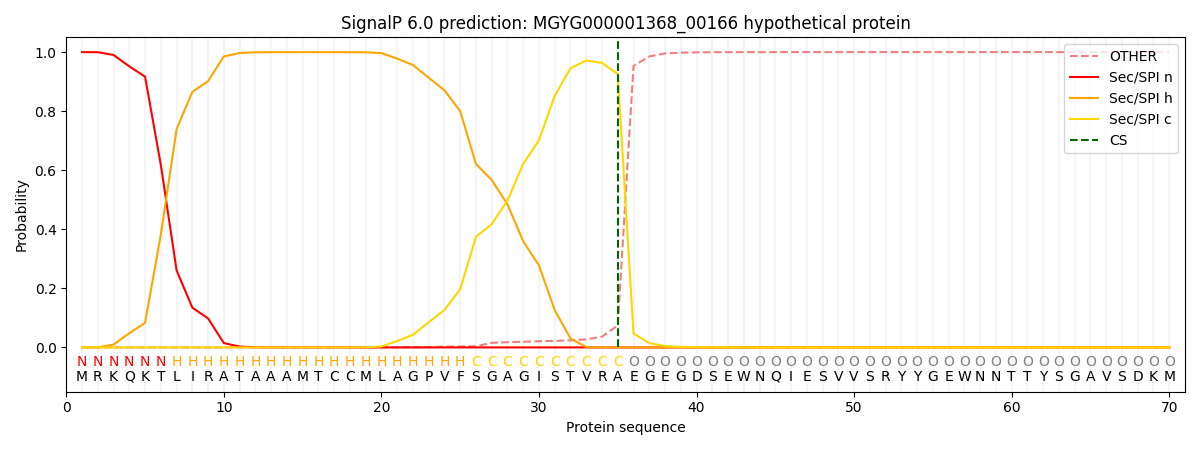
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000242 | 0.999081 | 0.000205 | 0.000179 | 0.000145 | 0.000126 |

