You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001368_03445
You are here: Home > Sequence: MGYG000001368_03445
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
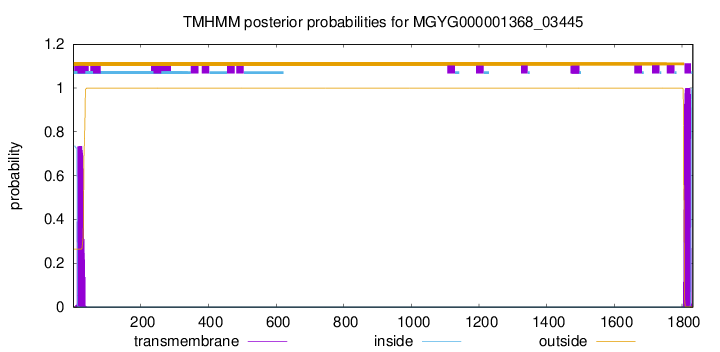
TMHMM annotations
Basic Information help
| Species | Muricomes fissicatena_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Muricomes; Muricomes fissicatena_A | |||||||||||
| CAZyme ID | MGYG000001368_03445 | |||||||||||
| CAZy Family | GH98 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 205940; End: 211438 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH98 | 971 | 1284 | 4.2e-20 | 0.8776758409785933 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08307 | Glyco_hydro_98C | 3.32e-20 | 1311 | 1555 | 1 | 269 | Glycosyl hydrolase family 98 C-terminal domain. This putative domain is found at the C-terminus of glycosyl hydrolase family 98 proteins. This domain is not expected to form part of the catalytic activity. |
| pfam08306 | Glyco_hydro_98M | 3.04e-12 | 1025 | 1306 | 29 | 328 | Glycosyl hydrolase family 98. This domain is the putative catalytic domain of glycosyl hydrolase family 98 proteins. |
| pfam02368 | Big_2 | 4.32e-05 | 875 | 962 | 2 | 76 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| pfam00754 | F5_F8_type_C | 5.05e-04 | 97 | 192 | 31 | 111 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIA30400.1 | 4.72e-116 | 478 | 966 | 22 | 503 |
| QQR05375.1 | 8.80e-116 | 478 | 966 | 22 | 503 |
| ANU41753.1 | 8.80e-116 | 478 | 966 | 22 | 503 |
| QIG40118.1 | 1.09e-83 | 967 | 1553 | 31 | 566 |
| VFH47827.1 | 8.01e-39 | 960 | 1558 | 25 | 594 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6N1B_A | 8.82e-120 | 475 | 966 | 13 | 497 | Crystalstructure of an N-acetylgalactosamine deacetylase from F. plautii in complex with blood group B trisaccharide [Flavonifractor plautii] |
| 6N1A_A | 2.06e-118 | 475 | 966 | 13 | 497 | Crystalstructure of an N-acetylgalactosamine deacetylase from F. plautii [Flavonifractor plautii] |
| 2WMH_A | 7.95e-40 | 949 | 1553 | 12 | 581 | Crystalstructure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae TIGR4 in complex with the H- disaccharide blood group antigen. [Streptococcus pneumoniae TIGR4] |
| 2WMF_A | 3.44e-39 | 949 | 1553 | 12 | 581 | Crystalstructure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae TIGR4 (Sp4GH98) in its native form. [Streptococcus pneumoniae TIGR4] |
| 2WMG_A | 8.29e-39 | 949 | 1553 | 12 | 581 | Crystalstructure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae TIGR4 (Sp4GH98) in complex with the LewisY pentasaccharide blood group antigen. [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0DTR4 | 1.76e-116 | 478 | 966 | 22 | 503 | A type blood N-acetyl-alpha-D-galactosamine deacetylase OS=Flavonifractor plautii OX=292800 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000911 | 0.998334 | 0.000206 | 0.000207 | 0.000164 | 0.000153 |