You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001370_01843
Basic Information
help
| Species |
Bacteroides fluxus
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides fluxus
|
| CAZyme ID |
MGYG000001370_01843
|
| CAZy Family |
CE19 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001370 |
4330086 |
Isolate |
not provided |
not provided |
|
| Gene Location |
Start: 65350;
End: 66504
Strand: +
|
No EC number prediction in MGYG000001370_01843.
| Family |
Start |
End |
Evalue |
family coverage |
| CE19 |
61 |
384 |
1.6e-25 |
0.8493975903614458 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG0412
|
DLH |
1.25e-17 |
110 |
337 |
13 |
178 |
Dienelactone hydrolase [Secondary metabolites biosynthesis, transport and catabolism]. |
| COG1506
|
DAP2 |
1.03e-09 |
91 |
384 |
358 |
618 |
Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| pfam01738
|
DLH |
3.51e-05 |
112 |
271 |
1 |
125 |
Dienelactone hydrolase family. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
O34973
|
1.23e-23 |
62 |
383 |
2 |
299 |
Putative hydrolase YtaP OS=Bacillus subtilis (strain 168) OX=224308 GN=ytaP PE=3 SV=1 |
This protein is predicted as SP
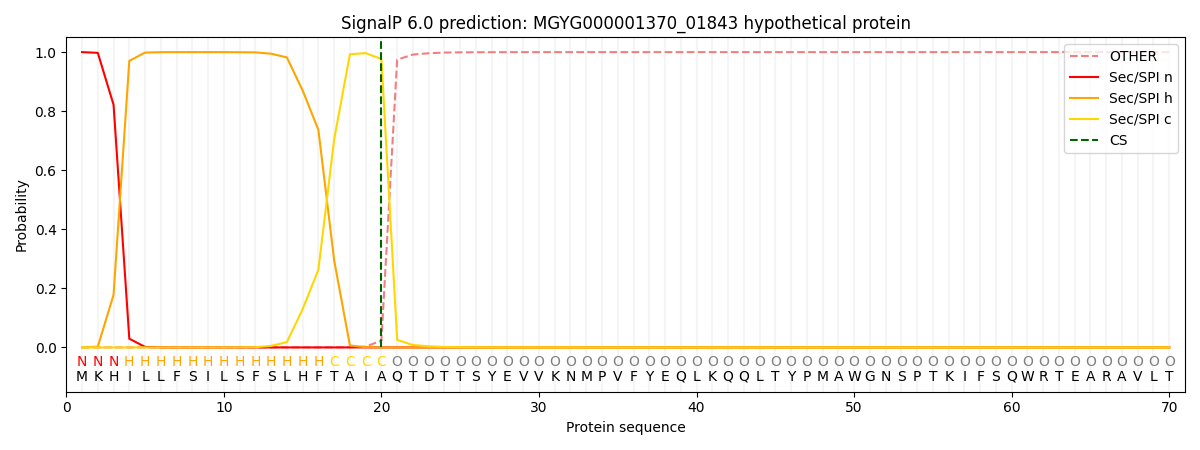
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000293
|
0.999052
|
0.000170
|
0.000160
|
0.000146
|
0.000135
|
There is no transmembrane helices in MGYG000001370_01843.

