You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001371_00589
You are here: Home > Sequence: MGYG000001371_00589
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus lautus_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus lautus_A | |||||||||||
| CAZyme ID | MGYG000001371_00589 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 64609; End: 67683 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 206 | 553 | 1.9e-43 | 0.9507692307692308 |
| CBM66 | 862 | 1015 | 1.1e-28 | 0.9806451612903225 |
| CBM66 | 577 | 734 | 1.7e-25 | 0.9806451612903225 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 3.56e-22 | 176 | 639 | 82 | 523 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| cd11304 | Cadherin_repeat | 5.34e-13 | 753 | 842 | 5 | 97 | Cadherin tandem repeat domain. Cadherins are glycoproteins involved in Ca2+-mediated cell-cell adhesion. The cadherin repeat domains occur as tandem repeats in the extracellular regions, which are thought to mediate cell-cell contact when bound to calcium. They play numerous roles in cell fate, signalling, proliferation, differentiation, and migration; members include E-, N-, P-, T-, VE-, CNR-, proto-, and FAT-family cadherin, desmocollin, and desmoglein, a large variety of domain architectures with varying repeat copy numbers. Cadherin-repeat containing proteins exist as monomers, homodimers, or heterodimers. |
| pfam06439 | DUF1080 | 8.23e-09 | 858 | 1015 | 6 | 178 | Domain of Unknown Function (DUF1080). This family has structural similarity to an endo-1,3-1,4-beta glucanase belonging to glycoside hydrolase family 16. However, the structure surrounding the active site differs from that of the endo-1,3-1,4-beta glucanase. |
| smart00112 | CA | 2.33e-08 | 768 | 844 | 1 | 80 | Cadherin repeats. Cadherins are glycoproteins involved in Ca2+-mediated cell-cell adhesion. Cadherin domains occur as repeats in the extracellular regions which are thought to mediate cell-cell contact when bound to calcium. |
| pfam06439 | DUF1080 | 1.84e-07 | 573 | 734 | 6 | 178 | Domain of Unknown Function (DUF1080). This family has structural similarity to an endo-1,3-1,4-beta glucanase belonging to glycoside hydrolase family 16. However, the structure surrounding the active site differs from that of the endo-1,3-1,4-beta glucanase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ACX65654.1 | 0.0 | 1 | 1024 | 1 | 1024 |
| QOT12100.1 | 0.0 | 1 | 1024 | 1 | 1024 |
| QUL54880.1 | 0.0 | 4 | 1018 | 9 | 1048 |
| CQR58237.1 | 4.46e-141 | 54 | 554 | 25 | 513 |
| QUL57088.1 | 2.51e-140 | 54 | 554 | 26 | 514 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2UVE_A | 5.70e-08 | 168 | 380 | 146 | 403 | Structureof Yersinia enterocolitica Family 28 Exopolygalacturonase [Yersinia enterocolitica],2UVE_B Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase [Yersinia enterocolitica],2UVF_A Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase in Complex with Digalaturonic Acid [Yersinia enterocolitica],2UVF_B Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase in Complex with Digalaturonic Acid [Yersinia enterocolitica] |
| 5OLP_A | 7.84e-08 | 198 | 472 | 64 | 355 | Galacturonidase[Bacteroides thetaiotaomicron VPI-5482],5OLP_B Galacturonidase [Bacteroides thetaiotaomicron VPI-5482] |
| 4MXN_A | 1.08e-07 | 190 | 292 | 33 | 127 | Crystalstructure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_B Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_C Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_D Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P15922 | 1.04e-07 | 176 | 556 | 151 | 573 | Exo-poly-alpha-D-galacturonosidase OS=Dickeya chrysanthemi OX=556 GN=pehX PE=1 SV=1 |
| O88277 | 5.41e-06 | 692 | 844 | 3268 | 3425 | Protocadherin Fat 2 OS=Rattus norvegicus OX=10116 GN=Fat2 PE=1 SV=1 |
| Q5F226 | 5.41e-06 | 692 | 844 | 3268 | 3425 | Protocadherin Fat 2 OS=Mus musculus OX=10090 GN=Fat2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
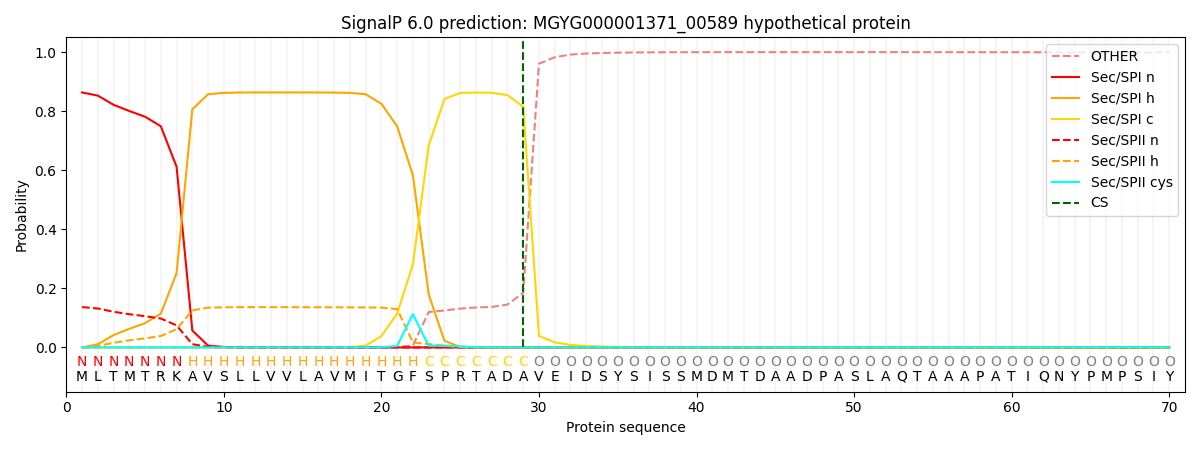
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000630 | 0.854574 | 0.143839 | 0.000339 | 0.000352 | 0.000249 |
