You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001371_01327
You are here: Home > Sequence: MGYG000001371_01327
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus lautus_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus lautus_A | |||||||||||
| CAZyme ID | MGYG000001371_01327 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 408; End: 1727 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL7 | 184 | 434 | 7e-85 | 0.9925093632958801 |
| CBM32 | 41 | 160 | 2.5e-22 | 0.9435483870967742 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08787 | Alginate_lyase2 | 1.91e-59 | 184 | 438 | 1 | 222 | Alginate lyase. Alginate lyases are enzymes that degrade the linear polysaccharide alignate. They cleave the glycosidic linkage of alignate through a beta-elimination reaction. This family forms an all beta fold and is different to all alpha fold of pfam05426. |
| pfam00754 | F5_F8_type_C | 1.08e-16 | 41 | 160 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 1.29e-06 | 39 | 152 | 11 | 134 | Substituted updates: Jan 31, 2002 |
| pfam17132 | Glyco_hydro_106 | 7.63e-04 | 54 | 145 | 184 | 283 | alpha-L-rhamnosidase. |
| smart00231 | FA58C | 0.004 | 53 | 153 | 22 | 130 | Coagulation factor 5/8 C-terminal domain, discoidin domain. Cell surface-attached carbohydrate-binding domain, present in eukaryotes and assumed to have horizontally transferred to eubacterial genomes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AYB42356.1 | 8.33e-311 | 1 | 439 | 1 | 439 |
| QOT10300.1 | 1.68e-310 | 1 | 439 | 1 | 439 |
| ACX67576.1 | 3.63e-305 | 1 | 439 | 1 | 439 |
| AFF61390.1 | 8.53e-304 | 1 | 439 | 1 | 439 |
| AKJ04963.1 | 5.11e-144 | 19 | 439 | 8 | 512 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5ZU5_A | 3.60e-70 | 39 | 438 | 34 | 508 | Crystalstructure of a full length alginate lyase with CBM domain [Vibrio splendidus] |
| 4OZX_A | 3.80e-53 | 180 | 438 | 3 | 281 | ChainA, Alginate lyase [Klebsiella pneumoniae] |
| 5XNR_A | 4.38e-37 | 59 | 246 | 28 | 217 | TruncatedAlyQ with CBM32 and alginate lyase domains [Persicobacter sp. CCB-QB2] |
| 7D29_A | 2.95e-28 | 59 | 171 | 28 | 140 | CBM32of AlyQ [Persicobacter sp. CCB-QB2],7D2A_A CBM32 of AlyQ in complex with 4,5-unsaturated mannuronic acid [Persicobacter sp. CCB-QB2] |
| 5ZU6_A | 3.37e-24 | 39 | 158 | 34 | 152 | ACBM32 derived from alginate lyase B (AlyB-OU02) [Vibrio] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q59478 | 3.83e-52 | 180 | 438 | 25 | 303 | Alginate lyase OS=Klebsiella pneumoniae OX=573 GN=alyA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
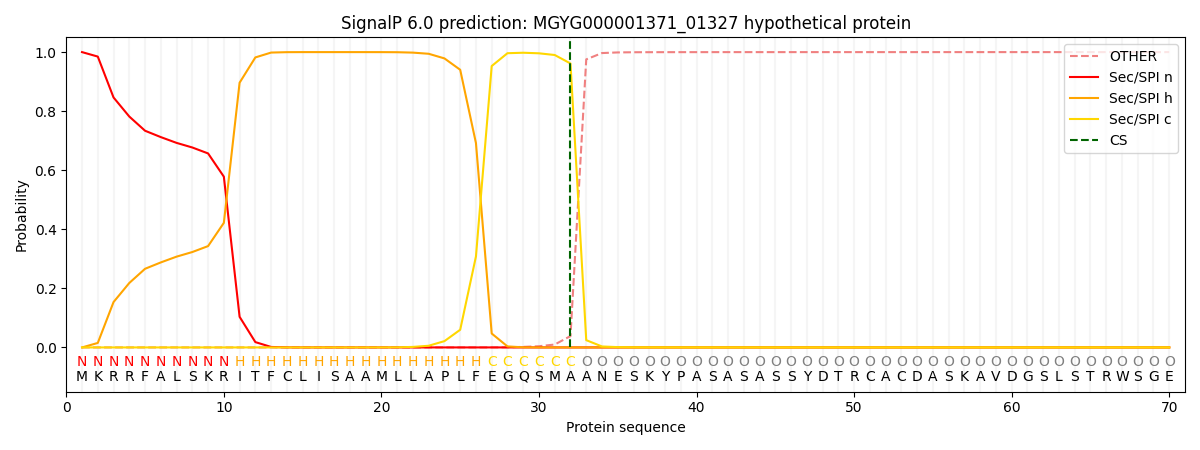
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000311 | 0.998964 | 0.000185 | 0.000203 | 0.000158 | 0.000143 |
