You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001371_01328
You are here: Home > Sequence: MGYG000001371_01328
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus lautus_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus lautus_A | |||||||||||
| CAZyme ID | MGYG000001371_01328 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | Acetyl esterase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1825; End: 6789 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 855 | 970 | 1.5e-19 | 0.9112903225806451 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0657 | Aes | 2.60e-40 | 272 | 524 | 71 | 302 | Acetyl esterase/lipase [Lipid transport and metabolism]. |
| pfam07859 | Abhydrolase_3 | 4.82e-22 | 283 | 506 | 1 | 207 | alpha/beta hydrolase fold. This catalytic domain is found in a very wide range of enzymes. |
| PRK10162 | PRK10162 | 5.47e-19 | 275 | 506 | 76 | 291 | acetyl esterase. |
| pfam00754 | F5_F8_type_C | 1.72e-17 | 852 | 969 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| NF033679 | DNRLRE_dom | 5.81e-16 | 573 | 739 | 1 | 163 | DNRLRE domain. The DNRLRE domain, with a length of about 160 amino acids, appears typically in large, repetitive surface proteins of bacteria and archaea, sometimes repeated several times. It occurs, notably, three times in the C-terminal region of the enzyme disaggregatase from the archaeal species Methanosarcina mazei, each time with the motif DNRLRE, for which the domain is named. Archaeal proteins within this family are described particularly well by the currently more narrowly defined Pfam model, PF06848. Note that the catalytic region of disaggregatase, in the N-terminal portion of the protein, is modeled by a different HMM, PF08480. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ACX67577.1 | 0.0 | 1 | 1654 | 1 | 1654 |
| AYB42355.1 | 0.0 | 1 | 1654 | 1 | 1654 |
| QOT10299.1 | 0.0 | 1 | 1654 | 1 | 1654 |
| APO44170.1 | 0.0 | 55 | 1449 | 54 | 1440 |
| QLG42471.1 | 0.0 | 53 | 1449 | 52 | 1440 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6MLY_A | 4.94e-32 | 282 | 505 | 548 | 769 | ChainA, Bifunctional GH43-CE protein [Bacteroides eggerthii],6MLY_B Chain B, Bifunctional GH43-CE protein [Bacteroides eggerthii],6MLY_C Chain C, Bifunctional GH43-CE protein [Bacteroides eggerthii],6MLY_D Chain D, Bifunctional GH43-CE protein [Bacteroides eggerthii] |
| 7D29_A | 5.84e-29 | 848 | 979 | 9 | 139 | CBM32of AlyQ [Persicobacter sp. CCB-QB2],7D2A_A CBM32 of AlyQ in complex with 4,5-unsaturated mannuronic acid [Persicobacter sp. CCB-QB2] |
| 5XNR_A | 1.87e-26 | 848 | 979 | 9 | 139 | TruncatedAlyQ with CBM32 and alginate lyase domains [Persicobacter sp. CCB-QB2] |
| 5ZU6_A | 1.67e-22 | 851 | 970 | 35 | 158 | ACBM32 derived from alginate lyase B (AlyB-OU02) [Vibrio] |
| 5ZU5_A | 4.09e-20 | 851 | 970 | 35 | 158 | Crystalstructure of a full length alginate lyase with CBM domain [Vibrio splendidus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q50681 | 3.01e-25 | 258 | 506 | 161 | 392 | Probable carboxylic ester hydrolase LipM OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=lipM PE=1 SV=1 |
| P96402 | 4.31e-22 | 278 | 506 | 154 | 369 | Esterase LipC OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=lipC PE=1 SV=1 |
| I6Y9F7 | 2.34e-21 | 254 | 506 | 142 | 379 | Esterase LipQ OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=lipQ PE=1 SV=1 |
| P9WK87 | 2.73e-15 | 253 | 518 | 55 | 305 | Carboxylesterase NlhH OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=nlhH PE=1 SV=1 |
| P9WK86 | 2.73e-15 | 253 | 518 | 55 | 305 | Carboxylesterase NlhH OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=nlhH PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
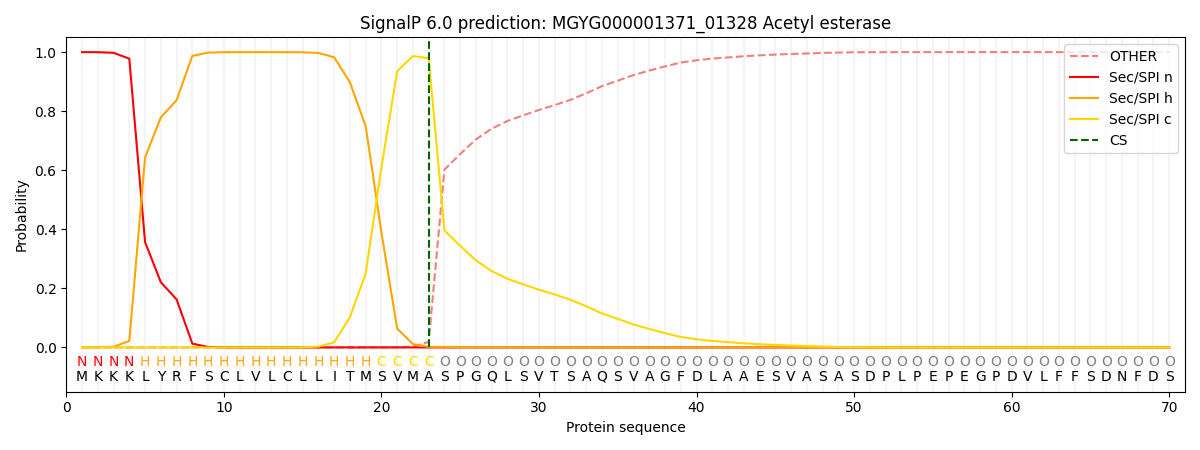
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000767 | 0.998105 | 0.000490 | 0.000209 | 0.000195 | 0.000201 |

