You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001371_01557
You are here: Home > Sequence: MGYG000001371_01557
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus lautus_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus lautus_A | |||||||||||
| CAZyme ID | MGYG000001371_01557 | |||||||||||
| CAZy Family | GH19 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 22373; End: 23647 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH19 | 192 | 417 | 1.1e-50 | 0.8658008658008658 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd00325 | chitinase_GH19 | 7.45e-66 | 212 | 418 | 35 | 224 | Glycoside hydrolase family 19, chitinase domain. Chitinases are enzymes that catalyze the hydrolysis of the beta-1,4-N-acetyl-D-glucosamine linkages in chitin polymers. Glycoside hydrolase family 19 chitinases are found primarily in plants (classes I, III, and IV), but some are found in bacteria. Class I and II chitinases are similar in their catalytic domains. Class I chitinases have an N-terminal cysteine-rich, chitin-binding domain which is separated from the catalytic domain by a proline and glycine-rich hinge region. Class II chitinases lack both the chitin-binding domain and the hinge region. Class IV chitinases are similar to class I chitinases, but they are smaller in size due to certain deletions. Despite lacking any significant sequence homology with lysozymes, structural analysis reveals that family 19 chitinases, together with family 46 chitosanases, are similar to several lysozymes including those from T4-phage and from goose. The structures reveal that the different enzyme groups arose from a common ancestor glycohydrolase antecedent to the prokaryotic/eukaryotic divergence. |
| pfam00182 | Glyco_hydro_19 | 7.54e-45 | 222 | 417 | 49 | 231 | Chitinase class I. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ACX63459.1 | 3.36e-307 | 1 | 424 | 1 | 424 |
| QOT08348.1 | 1.59e-305 | 1 | 424 | 1 | 424 |
| AYB46114.1 | 6.39e-295 | 1 | 424 | 1 | 420 |
| AVV54755.1 | 1.10e-267 | 1 | 424 | 1 | 427 |
| ANA81127.1 | 1.10e-267 | 1 | 424 | 1 | 427 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3CQL_A | 2.74e-45 | 222 | 424 | 53 | 242 | CrystalStructure of GH family 19 chitinase from Carica papaya [Carica papaya],3CQL_B Crystal Structure of GH family 19 chitinase from Carica papaya [Carica papaya] |
| 2DKV_A | 1.14e-44 | 210 | 424 | 101 | 298 | Crystalstructure of class I chitinase from Oryza sativa L. japonica [Oryza sativa Japonica Group],3IWR_A Crystal structure of class I chitinase from Oryza sativa L. japonica [Oryza sativa Japonica Group],3IWR_B Crystal structure of class I chitinase from Oryza sativa L. japonica [Oryza sativa Japonica Group] |
| 7V92_A | 2.19e-44 | 222 | 424 | 53 | 242 | ChainA, GH19 Chitinase [Ficus microcarpa],7V92_B Chain B, GH19 Chitinase [Ficus microcarpa],7V92_C Chain C, GH19 Chitinase [Ficus microcarpa],7V92_D Chain D, GH19 Chitinase [Ficus microcarpa] |
| 2BAA_A | 3.07e-43 | 222 | 424 | 53 | 242 | TheRefined Crystal Structure Of An Endochitinase From Hordeum Vulgare L. Seeds To 1.8 Angstroms Resolution [Hordeum vulgare] |
| 7V91_A | 3.24e-43 | 222 | 424 | 53 | 242 | ChainA, GH19 Chitinase [Ficus microcarpa],7V91_B Chain B, GH19 Chitinase [Ficus microcarpa],7V91_C Chain C, GH19 Chitinase [Ficus microcarpa],7V91_D Chain D, GH19 Chitinase [Ficus microcarpa] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q05537 | 7.33e-47 | 201 | 424 | 35 | 245 | Basic endochitinase (Fragment) OS=Solanum lycopersicum OX=4081 GN=CHI14 PE=2 SV=1 |
| P85084 | 1.50e-44 | 222 | 424 | 53 | 242 | Endochitinase OS=Carica papaya OX=3649 PE=1 SV=1 |
| Q688M5 | 1.56e-44 | 213 | 424 | 128 | 326 | Chitinase 9 OS=Oryza sativa subsp. japonica OX=39947 GN=Cht9 PE=2 SV=1 |
| P86473 | 3.17e-44 | 211 | 424 | 113 | 313 | Endochitinase OS=Actinidia chinensis var. chinensis OX=1590841 GN=CEY00_Acc04863 PE=1 SV=2 |
| Q5NB11 | 1.05e-43 | 213 | 424 | 91 | 290 | Chitinase 10 OS=Oryza sativa subsp. japonica OX=39947 GN=Cht10 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
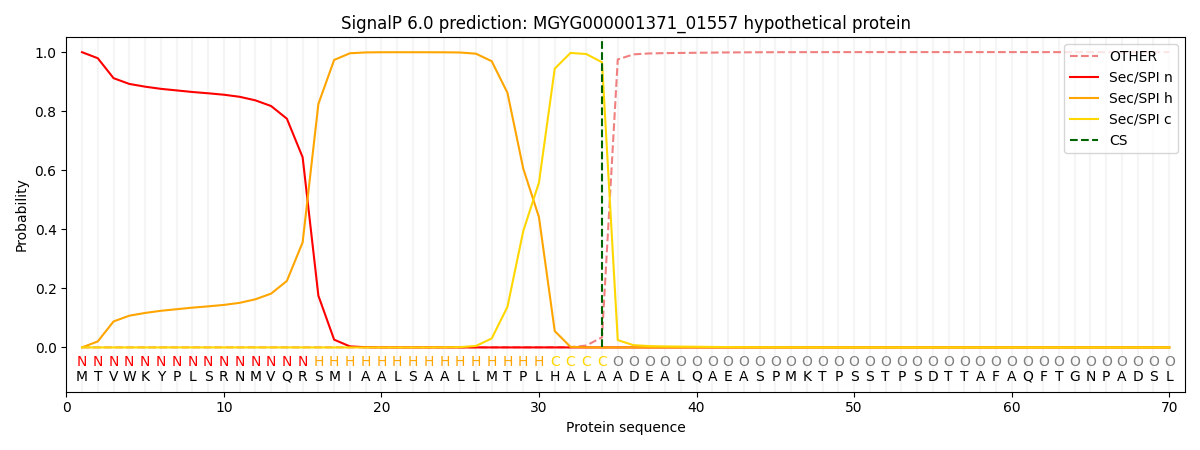
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001081 | 0.997779 | 0.000417 | 0.000326 | 0.000211 | 0.000167 |
