You are browsing environment: HUMAN GUT
MGYG000001371_01580
Basic Information
help
Species
Paenibacillus lautus_A
Lineage
Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus lautus_A
CAZyme ID
MGYG000001371_01580
CAZy Family
CBM32
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000001371
6950059
Isolate
not provided
not provided
Gene Location
Start: 48948;
End: 51728
Strand: -
No EC number prediction in MGYG000001371_01580.
Family
Start
End
Evalue
family coverage
CBM32
53
171
4.7e-18
0.9193548387096774
PL29
445
731
2.6e-16
0.9036544850498339
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam00754
F5_F8_type_C
2.19e-13
52
160
1
113
F5/8 type C domain. This domain is also known as the discoidin (DS) domain family.
more
cd00057
FA58C
9.12e-07
53
175
14
142
Substituted updates: Jan 31, 2002
more
pfam12708
Pectate_lyase_3
2.91e-06
457
573
16
134
Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28.
more
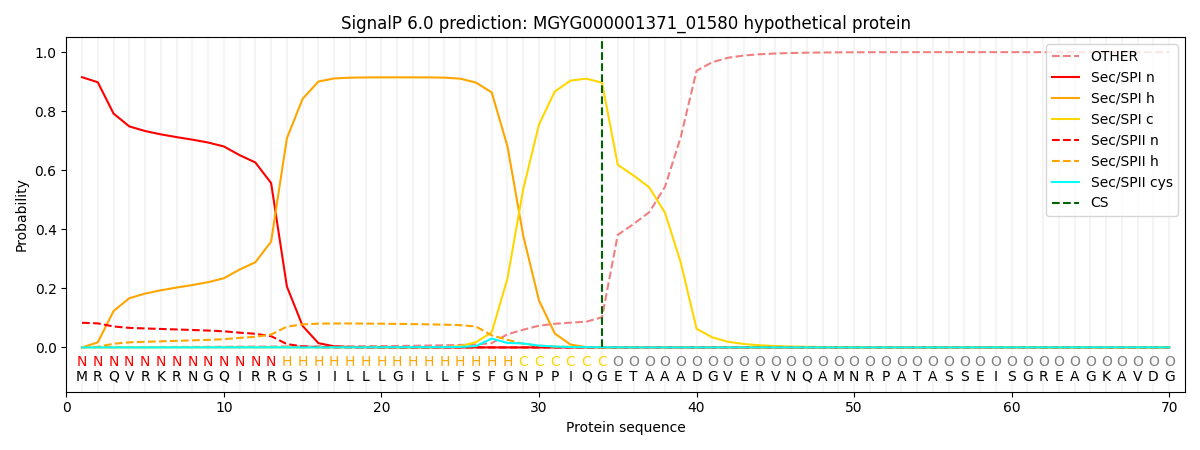
This protein is predicted as SP
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.002123
0.910938
0.086119
0.000365
0.000236
0.000198