You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001371_02618
You are here: Home > Sequence: MGYG000001371_02618
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
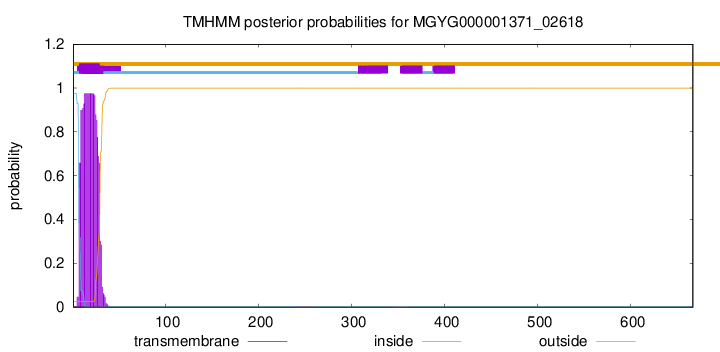
TMHMM annotations
Basic Information help
| Species | Paenibacillus lautus_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus lautus_A | |||||||||||
| CAZyme ID | MGYG000001371_02618 | |||||||||||
| CAZy Family | GH93 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 49344; End: 51347 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH93 | 44 | 326 | 1.9e-90 | 0.9022801302931596 |
| CBM42 | 538 | 665 | 5.1e-38 | 0.9779411764705882 |
| CBM13 | 391 | 535 | 4.6e-27 | 0.7287234042553191 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam05270 | AbfB | 5.42e-41 | 537 | 666 | 1 | 137 | Alpha-L-arabinofuranosidase B (ABFB) domain. This family consists of several fungal alpha-L-arabinofuranosidase B proteins. L-Arabinose is a constituent of plant-cell-wall poly-saccharides. It is found in a polymeric form in L-arabinan, in which the backbone is formed by 1,5-a- linked l-arabinose residues that can be branched via 1,2-a- and 1,3-a-linked l-arabinofuranose side chains. AbfB hydrolyzes 1,5-a, 1,3-a and 1,2-a linkages in both oligosaccharides and polysaccharides, which contain terminal non-reducing l-arabinofuranoses in side chains. |
| pfam14200 | RicinB_lectin_2 | 1.77e-18 | 385 | 468 | 6 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 4.92e-18 | 427 | 515 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam00652 | Ricin_B_lectin | 9.47e-15 | 393 | 525 | 2 | 126 | Ricin-type beta-trefoil lectin domain. |
| cd15482 | Sialidase_non-viral | 5.06e-11 | 54 | 215 | 86 | 239 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOT10786.1 | 0.0 | 1 | 667 | 1 | 667 |
| ACX67045.1 | 0.0 | 1 | 667 | 1 | 667 |
| SDS04433.1 | 0.0 | 1 | 667 | 1 | 667 |
| CQR58317.1 | 1.11e-306 | 5 | 667 | 1 | 662 |
| QJD84758.1 | 2.06e-303 | 1 | 667 | 1 | 667 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3A71_A | 1.44e-29 | 36 | 335 | 11 | 309 | Highresolution structure of Penicillium chrysogenum alpha-L-arabinanase [Penicillium chrysogenum],3A72_A High resolution structure of Penicillium chrysogenum alpha-L-arabinanase complexed with arabinobiose [Penicillium chrysogenum] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
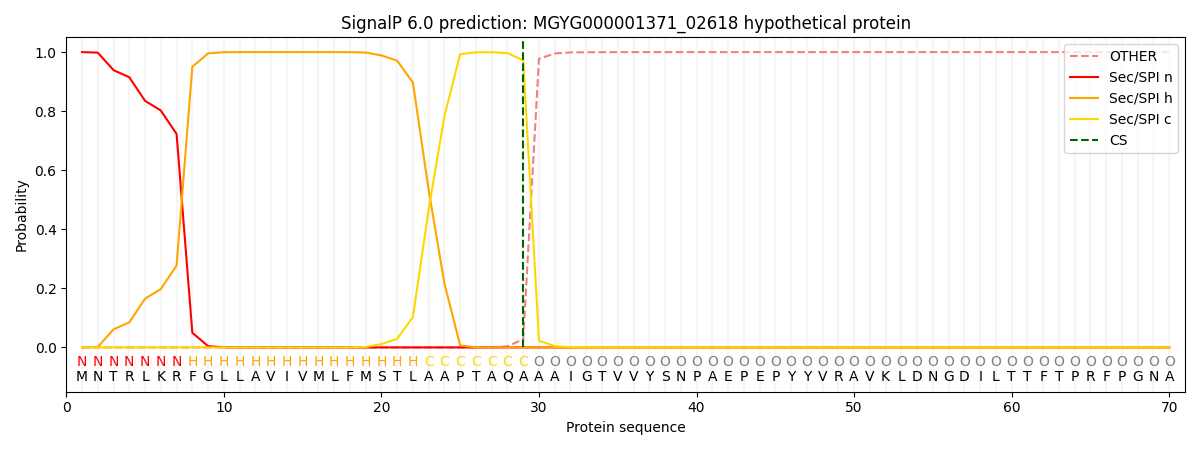
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000274 | 0.999011 | 0.000184 | 0.000202 | 0.000161 | 0.000143 |