You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001372_00327
You are here: Home > Sequence: MGYG000001372_00327
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paraprevotella xylaniphila | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Paraprevotella; Paraprevotella xylaniphila | |||||||||||
| CAZyme ID | MGYG000001372_00327 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 100168; End: 103431 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 45 | 504 | 9.9e-104 | 0.526595744680851 |
| GH43 | 837 | 1071 | 4.2e-42 | 0.9737991266375546 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd08983 | GH43_Bt3655-like | 2.42e-59 | 819 | 1086 | 1 | 262 | Glycosyl hydrolase family 43 protein such as Bacteroides thetaiotaomicron VPI-5482 arabinofuranosidase Bt3655. This glycosyl hydrolase family 43 (GH43)-like family includes the characterized arabinofuranosidases (EC 3.2.1.55): Bacteroides thetaiotaomicron VPI-5482 (Bt3655;BT_3655) and Penicillium chrysogenum 31B Abf43B, as well as Bifidobacterium adolescentis ATCC 15703 beta-xylosidase (EC 3.2.1.37) BAD_1527. It belongs to the glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) which includes family 43 (GH43) and 62 (GH62) families. GH43 includes enzymes with beta-xylosidase (EC 3.2.1.37), beta-1,3-xylosidase (EC 3.2.1.-), alpha-L-arabinofuranosidase (EC 3.2.1.55), arabinanase (EC 3.2.1.99), xylanase (EC 3.2.1.8), endo-alpha-L-arabinanases (beta-xylanases) and galactan 1,3-beta-galactosidase (EC 3.2.1.145) activities. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| COG3250 | LacZ | 4.55e-37 | 46 | 477 | 1 | 429 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 4.68e-27 | 118 | 448 | 69 | 417 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 2.43e-20 | 34 | 492 | 68 | 486 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam00703 | Glyco_hydro_2 | 1.93e-13 | 224 | 317 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT74822.1 | 1.52e-248 | 26 | 772 | 461 | 1205 |
| AGA25352.1 | 8.57e-213 | 23 | 762 | 24 | 749 |
| AYL99048.1 | 2.27e-208 | 10 | 762 | 6 | 734 |
| QNN25331.1 | 6.71e-208 | 9 | 772 | 5 | 754 |
| QEM14171.1 | 9.01e-208 | 10 | 762 | 6 | 734 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7SF2_A | 3.92e-151 | 24 | 608 | 3 | 586 | ChainA, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_B Chain B, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_C Chain C, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_D Chain D, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_E Chain E, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_F Chain F, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838] |
| 6XXW_A | 5.80e-24 | 50 | 477 | 23 | 444 | Structureof beta-D-Glucuronidase for Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12] |
| 6U7J_A | 9.98e-24 | 56 | 477 | 17 | 445 | UnculturedClostridium sp. Beta-glucuronidase [uncultured Clostridium sp.],6U7J_B Uncultured Clostridium sp. Beta-glucuronidase [uncultured Clostridium sp.],6U7J_C Uncultured Clostridium sp. Beta-glucuronidase [uncultured Clostridium sp.],6U7J_D Uncultured Clostridium sp. Beta-glucuronidase [uncultured Clostridium sp.] |
| 5Z1A_A | 9.14e-22 | 51 | 577 | 16 | 511 | Thecrystal structure of Bacteroides fragilis beta-glucuronidase in complex with uronic isofagomine [Bacteroides fragilis NCTC 9343] |
| 6ED2_A | 1.81e-21 | 50 | 448 | 30 | 441 | ChainA, Glycosyl hydrolase family 2, TIM barrel domain protein [Faecalibacterium duncaniae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KPJ7 | 1.09e-25 | 47 | 448 | 40 | 430 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_21970 PE=2 SV=1 |
| P77989 | 6.96e-23 | 109 | 448 | 51 | 388 | Beta-galactosidase OS=Thermoanaerobacter pseudethanolicus (strain ATCC 33223 / 39E) OX=340099 GN=lacZ PE=3 SV=2 |
| T2KM09 | 1.29e-17 | 118 | 482 | 110 | 459 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22050 PE=2 SV=2 |
| T2KN75 | 5.03e-15 | 62 | 504 | 30 | 499 | Beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22060 PE=1 SV=1 |
| A1SWB8 | 5.18e-15 | 115 | 492 | 119 | 498 | Beta-galactosidase OS=Psychromonas ingrahamii (strain 37) OX=357804 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
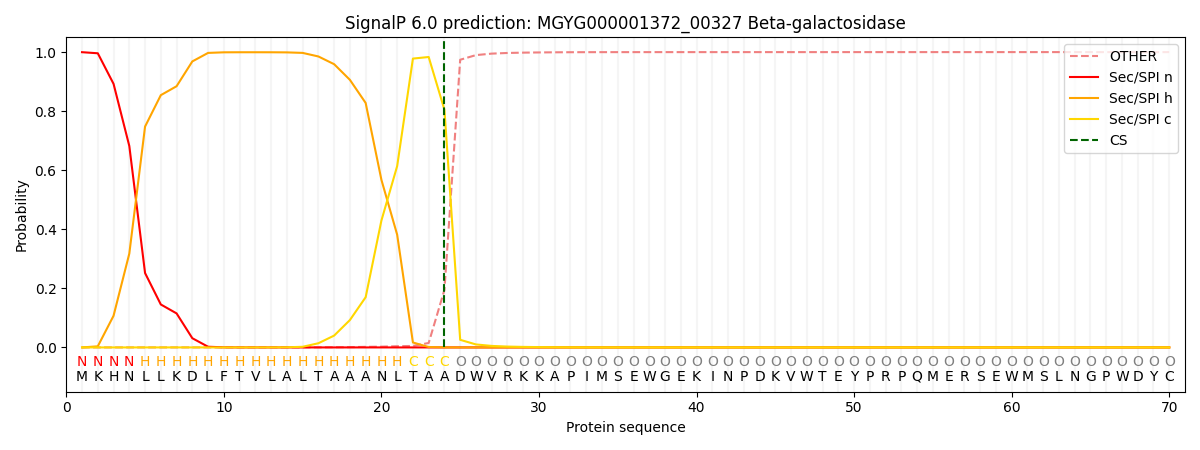
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000769 | 0.998138 | 0.000501 | 0.000221 | 0.000175 | 0.000162 |
