You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001372_02232
You are here: Home > Sequence: MGYG000001372_02232
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paraprevotella xylaniphila | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Paraprevotella; Paraprevotella xylaniphila | |||||||||||
| CAZyme ID | MGYG000001372_02232 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 61987; End: 63438 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 223 | 469 | 3.9e-44 | 0.7261538461538461 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 7.19e-12 | 193 | 368 | 196 | 366 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| PLN02218 | PLN02218 | 2.20e-10 | 246 | 467 | 200 | 410 | polygalacturonase ADPG |
| pfam00295 | Glyco_hydro_28 | 2.05e-08 | 241 | 435 | 88 | 276 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| PLN02793 | PLN02793 | 9.06e-07 | 247 | 436 | 186 | 368 | Probable polygalacturonase |
| PLN03003 | PLN03003 | 1.69e-05 | 251 | 400 | 151 | 291 | Probable polygalacturonase At3g15720 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUU00181.1 | 1.50e-153 | 4 | 483 | 5 | 459 |
| QUT92200.1 | 3.86e-153 | 7 | 477 | 6 | 450 |
| ALJ62050.1 | 5.46e-153 | 7 | 477 | 6 | 450 |
| QUR43693.1 | 2.41e-151 | 5 | 467 | 11 | 449 |
| QQR19335.1 | 4.83e-151 | 5 | 467 | 11 | 449 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3JUR_A | 2.47e-07 | 227 | 451 | 180 | 366 | Thecrystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_B The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_C The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_D The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B0Y9F8 | 5.30e-07 | 243 | 424 | 156 | 337 | Probable endo-xylogalacturonan hydrolase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=xghA PE=3 SV=2 |
| A1DBT5 | 2.17e-06 | 243 | 424 | 156 | 337 | Probable endo-xylogalacturonan hydrolase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=xghA PE=3 SV=1 |
| Q4WBT4 | 2.17e-06 | 243 | 424 | 156 | 337 | Probable endo-xylogalacturonan hydrolase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=xghA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
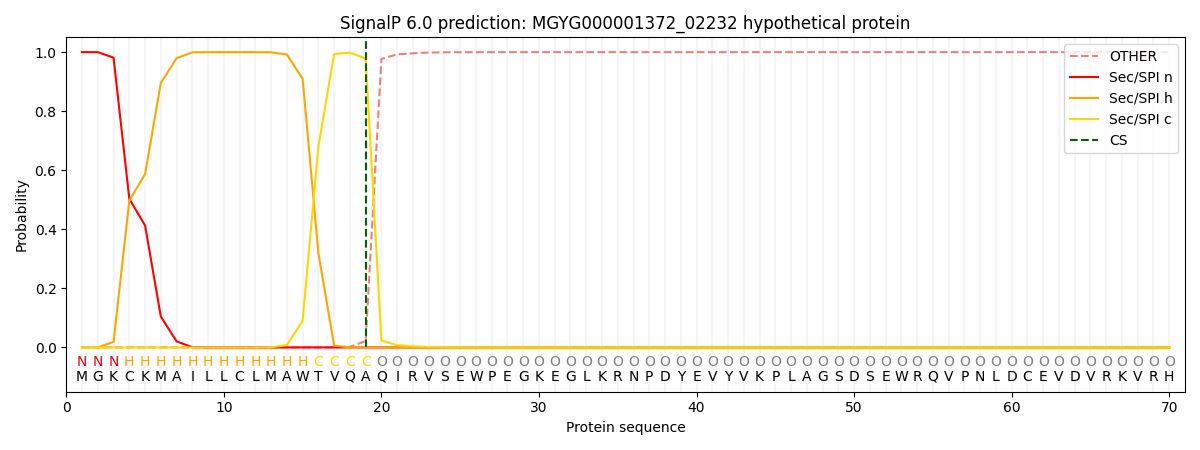
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000216 | 0.999158 | 0.000156 | 0.000149 | 0.000134 | 0.000130 |
