You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001373_00213
You are here: Home > Sequence: MGYG000001373_00213
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Faecalimonas sp000209385 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Faecalimonas; Faecalimonas sp000209385 | |||||||||||
| CAZyme ID | MGYG000001373_00213 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 225387; End: 228602 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 46 | 192 | 1.1e-24 | 0.9850746268656716 |
| CBM32 | 937 | 1046 | 3.6e-18 | 0.8064516129032258 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08305 | NPCBM | 3.86e-23 | 47 | 193 | 4 | 135 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 5.65e-20 | 47 | 193 | 6 | 144 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| pfam00754 | F5_F8_type_C | 7.02e-15 | 937 | 1052 | 4 | 120 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 3.01e-04 | 937 | 1012 | 17 | 90 | Substituted updates: Jan 31, 2002 |
| pfam13285 | DUF4073 | 0.005 | 840 | 897 | 86 | 139 | Domain of unknown function (DUF4073). This family is frequently found at the C-terminus of bacterial proteins carrying the family, Metallophos pfam00149. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AMN35280.1 | 5.79e-181 | 1 | 1071 | 1 | 991 |
| AQW23377.1 | 6.79e-179 | 31 | 1071 | 34 | 991 |
| ATD49073.1 | 7.80e-179 | 31 | 1071 | 39 | 996 |
| BCL58565.1 | 1.38e-147 | 58 | 1071 | 97 | 1052 |
| QTR94629.1 | 7.39e-104 | 47 | 1068 | 144 | 1099 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2V72_A | 4.50e-09 | 937 | 1031 | 19 | 124 | Thestructure of the family 32 CBM from C. perfringens NanJ in complex with galactose [Clostridium perfringens] |
| 4LKS_A | 5.50e-09 | 948 | 1066 | 41 | 165 | Structureof CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with galactose [Clostridium perfringens ATCC 13124],4LKS_C Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with galactose [Clostridium perfringens ATCC 13124],4LQR_A Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens ATCC 13124],4P5Y_A Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with N-acetylgalactosamine [Clostridium perfringens ATCC 13124] |
| 2KD7_A | 6.35e-09 | 943 | 1042 | 26 | 126 | ChainA, Putative chitobiase [Bacteroides thetaiotaomicron VPI-5482] |
| 3GGL_A | 7.77e-09 | 943 | 1042 | 36 | 136 | X-RayStructure of the C-terminal domain (277-440) of Putative chitobiase from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR324A. [Bacteroides thetaiotaomicron],6OE2_A X-Ray Structure of the C-terminal domain (277-440) of Putative chitobiase from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR324A. Re-refinement of 3GGL with correct metal Mn replacing Zn. New metal confirmed with PIXE analysis of original sample. [Bacteroides thetaiotaomicron] |
| 2VMH_A | 1.35e-08 | 47 | 191 | 10 | 148 | Thestructure of CBM51 from Clostridium perfringens GH95 [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0TR53 | 2.09e-06 | 937 | 1064 | 634 | 764 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| Q8XL08 | 2.09e-06 | 937 | 1064 | 634 | 764 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
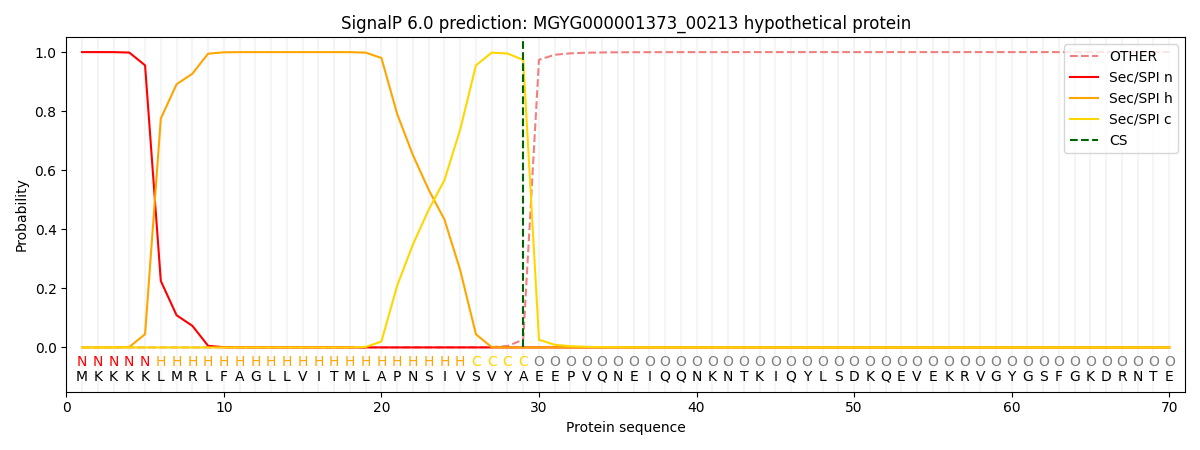
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000255 | 0.999064 | 0.000197 | 0.000160 | 0.000155 | 0.000141 |
