You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001374_00546
You are here: Home > Sequence: MGYG000001374_00546
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
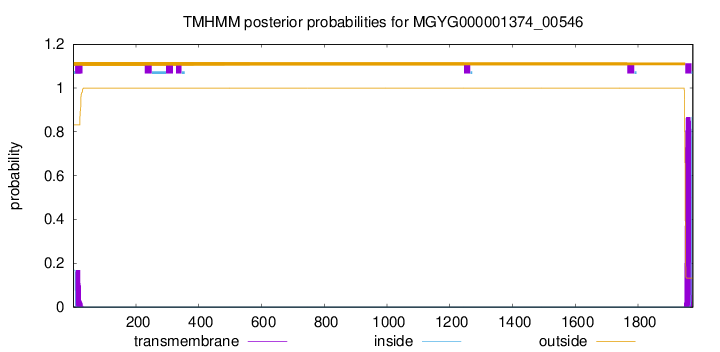
TMHMM annotations
Basic Information help
| Species | Mediterraneibacter torques | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Mediterraneibacter; Mediterraneibacter torques | |||||||||||
| CAZyme ID | MGYG000001374_00546 | |||||||||||
| CAZy Family | CE3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 582171; End: 588098 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH33 | 1202 | 1657 | 4e-82 | 0.9444444444444444 |
| CBM40 | 1021 | 1190 | 2e-41 | 0.8938547486033519 |
| CE3 | 632 | 785 | 2.3e-17 | 0.7835051546391752 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4409 | NanH | 1.70e-94 | 1035 | 1667 | 102 | 718 | Neuraminidase (sialidase) [Carbohydrate transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| cd15482 | Sialidase_non-viral | 5.60e-79 | 1199 | 1653 | 1 | 327 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| cd01834 | SGNH_hydrolase_like_2 | 4.51e-29 | 599 | 787 | 4 | 191 | SGNH_hydrolase subfamily. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| pfam13472 | Lipase_GDSL_2 | 6.68e-29 | 601 | 777 | 1 | 174 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| cd00229 | SGNH_hydrolase | 9.79e-28 | 599 | 786 | 1 | 186 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASM68050.1 | 2.80e-206 | 988 | 1677 | 38 | 724 |
| CAA69951.1 | 3.06e-206 | 998 | 1675 | 93 | 767 |
| ETD19277.1 | 1.18e-205 | 1003 | 1675 | 50 | 719 |
| QEI30999.1 | 1.18e-205 | 1003 | 1675 | 50 | 719 |
| QHB23506.1 | 1.18e-205 | 1003 | 1675 | 50 | 719 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1SLI_A | 3.96e-205 | 998 | 1678 | 1 | 678 | LeechIntramolecular Trans-Sialidase Complexed With Dana [Macrobdella decora],1SLL_A Sialidase L From Leech Macrobdella Decora [Macrobdella decora],2SLI_A Leech Intramolecular Trans-Sialidase Complexed With 2,7- Anhydro-Neu5ac, The Reaction Product [Macrobdella decora],3SLI_A Leech Intramolecular Trans-Sialidase Complexed With 2,7- Anhydro-Neu5ac Prepared By Soaking With 3'-Sialyllactose [Macrobdella decora],4SLI_A Leech Intramolecular Trans-Sialidase Complexed With 2- Propenyl-Neu5ac, An Inactive Substrate Analogue [Macrobdella decora] |
| 4X6K_A | 9.86e-153 | 1201 | 1675 | 7 | 482 | Crystalstructure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with Siastatin B [[Ruminococcus] gnavus CC55_001C] |
| 4X47_A | 1.09e-152 | 1201 | 1675 | 10 | 485 | Crystalstructure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with Neu5Ac2en [[Ruminococcus] gnavus ATCC 29149],4X49_A Crystal structure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with oseltamivir carboxylate [[Ruminococcus] gnavus ATCC 29149],4X4A_A Crystal structure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with 2,7-Anhydro-Neu5Ac [[Ruminococcus] gnavus ATCC 29149] |
| 4YW1_A | 7.14e-136 | 1007 | 1675 | 6 | 655 | ChainA, Neuraminidase C [Streptococcus pneumoniae TIGR4],4YW1_B Chain B, Neuraminidase C [Streptococcus pneumoniae TIGR4],4YW2_A Chain A, Neuraminidase C [Streptococcus pneumoniae TIGR4],4YW2_B Chain B, Neuraminidase C [Streptococcus pneumoniae TIGR4],4YW3_A Chain A, Neuraminidase C [Streptococcus pneumoniae TIGR4],4YW3_B Chain B, Neuraminidase C [Streptococcus pneumoniae TIGR4],4YW4_A Streptococcus pneumoniae sialidase NanC [Streptococcus pneumoniae],4YW4_B Streptococcus pneumoniae sialidase NanC [Streptococcus pneumoniae],4YW5_A Chain A, Neuraminidase C [Streptococcus pneumoniae TIGR4],4YW5_B Chain B, Neuraminidase C [Streptococcus pneumoniae TIGR4],5F9T_A Chain A, Neuraminidase C [Streptococcus pneumoniae TIGR4],5F9T_B Chain B, Neuraminidase C [Streptococcus pneumoniae TIGR4] |
| 4YZ1_A | 1.22e-135 | 1007 | 1675 | 25 | 674 | CrystalStructure of Streptococcus pneumoniae NanC, apo structure. [Streptococcus pneumoniae TIGR4],4YZ1_B Crystal Structure of Streptococcus pneumoniae NanC, apo structure. [Streptococcus pneumoniae TIGR4],4YZ2_A Crystal Structure of Streptococcus pneumoniae NanC, in complex with 2-deoxy-2,3-didehydro-N-acetylneuraminic acid. [Streptococcus pneumoniae],4YZ2_B Crystal Structure of Streptococcus pneumoniae NanC, in complex with 2-deoxy-2,3-didehydro-N-acetylneuraminic acid. [Streptococcus pneumoniae],4YZ3_A Crystal Structure of Streptococcus pneumoniae NanC, in complex with Oseltamivir. [Streptococcus pneumoniae TIGR4],4YZ3_B Crystal Structure of Streptococcus pneumoniae NanC, in complex with Oseltamivir. [Streptococcus pneumoniae TIGR4],4YZ4_A Crystal Structure of Streptococcus pneumoniae NanC, in complex with N-Acetylneuraminic acid. [Streptococcus pneumoniae],4YZ4_B Crystal Structure of Streptococcus pneumoniae NanC, in complex with N-Acetylneuraminic acid. [Streptococcus pneumoniae],4YZ5_A Crystal Structure of Streptococcus pneumoniae NanC, in complex with 3-Sialyllactose [Streptococcus pneumoniae],4YZ5_B Crystal Structure of Streptococcus pneumoniae NanC, in complex with 3-Sialyllactose [Streptococcus pneumoniae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q27701 | 2.46e-203 | 997 | 1678 | 80 | 758 | Anhydrosialidase OS=Macrobdella decora OX=6405 PE=1 SV=1 |
| Q54727 | 1.14e-130 | 1032 | 1675 | 68 | 695 | Sialidase B OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=nanB PE=1 SV=2 |
| P29767 | 1.87e-107 | 1031 | 1707 | 219 | 859 | Sialidase OS=Clostridium septicum OX=1504 PE=3 SV=1 |
| P62575 | 1.47e-56 | 1003 | 1649 | 121 | 768 | Sialidase A OS=Streptococcus pneumoniae OX=1313 GN=nanA PE=1 SV=1 |
| P62576 | 1.47e-56 | 1003 | 1649 | 121 | 768 | Sialidase A OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=nanA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
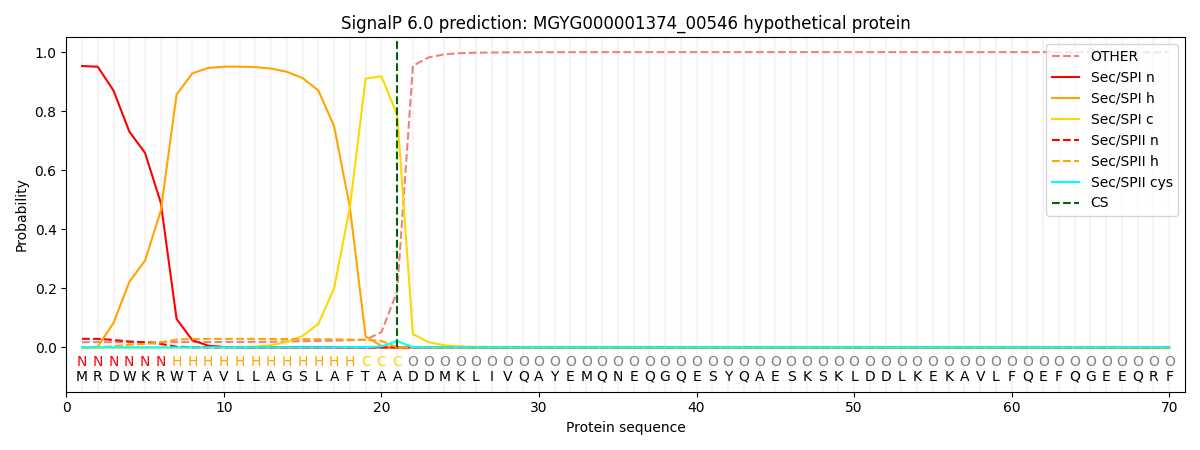
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.024716 | 0.943327 | 0.030950 | 0.000305 | 0.000327 | 0.000326 |