You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001374_02418
You are here: Home > Sequence: MGYG000001374_02418
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Mediterraneibacter torques | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Mediterraneibacter; Mediterraneibacter torques | |||||||||||
| CAZyme ID | MGYG000001374_02418 | |||||||||||
| CAZy Family | GH36 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 307006; End: 312177 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5492 | YjdB | 2.20e-17 | 1392 | 1560 | 182 | 324 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| COG5492 | YjdB | 6.85e-16 | 1467 | 1666 | 157 | 327 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| pfam00754 | F5_F8_type_C | 1.97e-12 | 142 | 265 | 6 | 114 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam02368 | Big_2 | 5.62e-12 | 1392 | 1467 | 1 | 76 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| pfam02368 | Big_2 | 8.31e-12 | 1492 | 1567 | 1 | 76 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBK22244.1 | 0.0 | 1 | 1374 | 1 | 1379 |
| BBK62317.1 | 0.0 | 1 | 1374 | 1 | 1379 |
| AMN35790.1 | 2.00e-217 | 14 | 1273 | 17 | 1287 |
| ASY51537.1 | 1.08e-215 | 14 | 1273 | 17 | 1287 |
| AWS26049.1 | 1.08e-215 | 14 | 1273 | 17 | 1287 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P33747 | 4.19e-09 | 1392 | 1560 | 38 | 190 | Uncharacterized protein CA_P0160 OS=Clostridium acetobutylicum (strain ATCC 824 / DSM 792 / JCM 1419 / LMG 5710 / VKM B-1787) OX=272562 GN=CA_P0160 PE=3 SV=2 |
| P35838 | 1.59e-06 | 1490 | 1576 | 235 | 321 | Uncharacterized protein CA_C0552 OS=Clostridium acetobutylicum (strain ATCC 824 / DSM 792 / JCM 1419 / LMG 5710 / VKM B-1787) OX=272562 GN=CA_C0552 PE=4 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
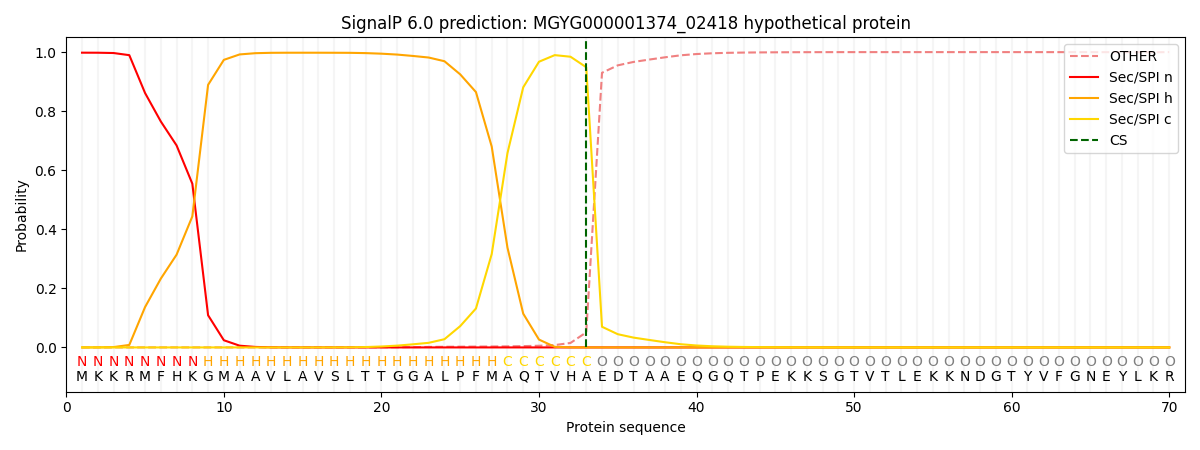
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000828 | 0.995741 | 0.002154 | 0.000747 | 0.000281 | 0.000208 |

