You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001375_00533
You are here: Home > Sequence: MGYG000001375_00533
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
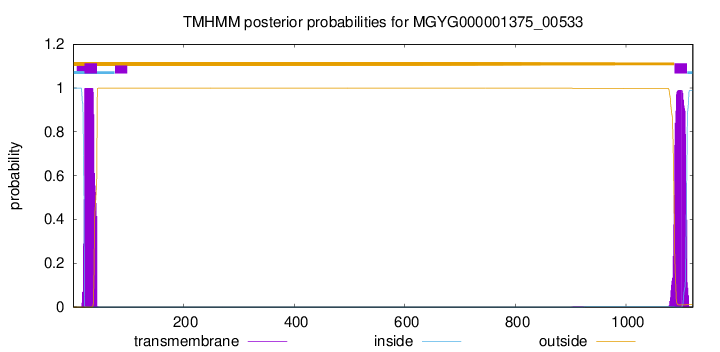
TMHMM annotations
Basic Information help
| Species | Ruminococcus_F champanellensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus_F; Ruminococcus_F champanellensis | |||||||||||
| CAZyme ID | MGYG000001375_00533 | |||||||||||
| CAZy Family | CBM22 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 591718; End: 595083 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM22 | 796 | 941 | 2.3e-23 | 0.9465648854961832 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam12892 | FctA | 4.04e-16 | 448 | 562 | 1 | 111 | Spy0128-like isopeptide containing domain. The FCT and equivalent region genes of Streptococcus pyogenes and other related bacteria encode surface proteins that include fibronectin- and collagen-binding proteins and the serological markers known as T antigens. Some of these proteins give rise to pilus-like appendages. The FctA family is found in many Firmicutes and related bacteria. In S. pyogenes, the pili have a role in bacterial adherence and colonisation of human tissues. Members of this family have a conserved N-terminal lysine and C-terminal asparagine that can form a covalent isopeptide bond. |
| pfam02018 | CBM_4_9 | 1.54e-09 | 796 | 935 | 3 | 120 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
| cd00222 | CollagenBindB | 4.58e-09 | 571 | 671 | 3 | 91 | Repeat unit of collagen-binding protein domain B. The collagen-binding protein mediates bacterial adherence to collagen; the primary sequence has a non-repetitive, collagen-binding A region, followed by instances of this B region repetitive unit. The B region has one to four 23 kDa repeat units (B1-B4), which have been suggested to serve as 'stalks' that project the A region from the bacterial surface and thus facilitate bacterial adherence to collagen. Each B repeat unit has two highly similar domains (D1 and D2) placed side-by-side; both D1 and D2 are included in this model. They exhibit a unique inverse IgG-like domain fold. |
| TIGR03786 | strep_pil_rpt | 1.40e-08 | 489 | 559 | 1 | 62 | streptococcal pilin isopeptide linkage domain. This model describes a domain that occurs once in the major pilin of Streptococcus pyogenes, Spy0128, but in higher copy numbers in other streptococcal proteins. The domain occurs nine times in a surface-anchored protein of Bifidobacterium longum. All members of this family have LPXTG-type sortase target sequences. The S. pyogenes major pilin has been shown to undergo isopeptide bond cross-linking, mediated by sortases, that are critical to maintaining pilus structural integrity. One such Lys-to-Asn isopeptide bond is to a near-invariant Asn near the C-terminal end of this domain (column 81 of the seed alignment). A Glu in the S. pyogenes major pilin (column 25 of the seed alignment), invariant as Glu or Gln, is described as catalytic for isopeptide bond formation. |
| pfam05738 | Cna_B | 1.95e-08 | 571 | 671 | 1 | 87 | Cna protein B-type domain. This domain is found in Staphylococcus aureus collagen-binding surface protein. The structure of the repetitive B-region has been solved and forms a beta sandwich structure. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL16766.1 | 0.0 | 1 | 1121 | 1 | 1121 |
| CBL18305.1 | 3.12e-16 | 801 | 980 | 498 | 652 |
| CAA84537.1 | 3.89e-14 | 768 | 960 | 230 | 397 |
| AEE64771.1 | 7.58e-14 | 801 | 1059 | 279 | 502 |
| AAT48119.1 | 7.62e-14 | 801 | 1059 | 281 | 504 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
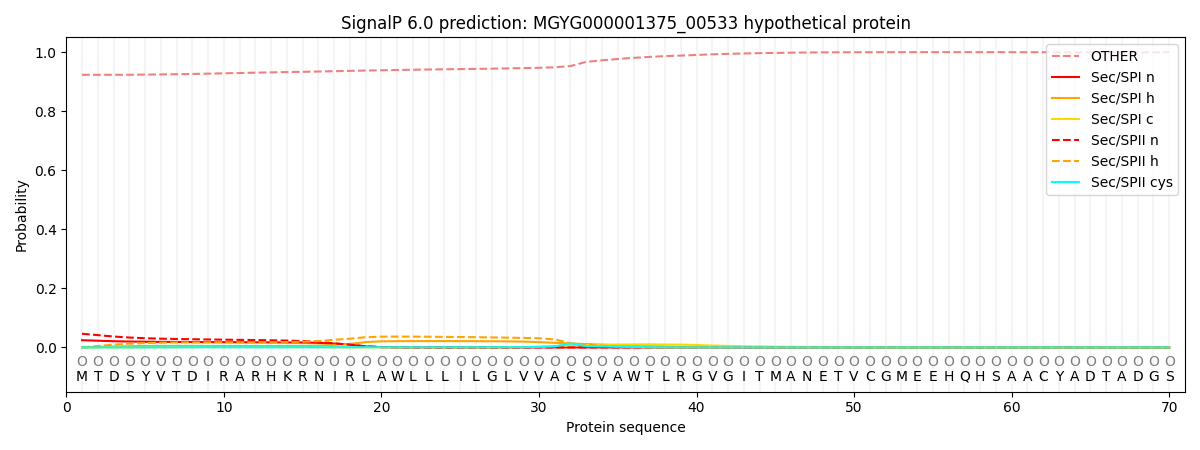
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.924259 | 0.022386 | 0.046572 | 0.000210 | 0.000109 | 0.006482 |