You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001375_02110
You are here: Home > Sequence: MGYG000001375_02110
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
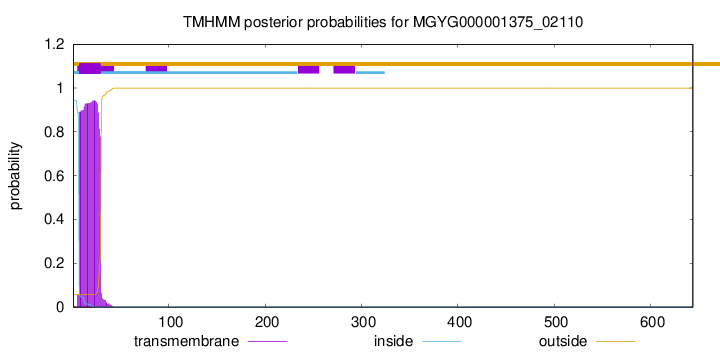
TMHMM annotations
Basic Information help
| Species | Ruminococcus_F champanellensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus_F; Ruminococcus_F champanellensis | |||||||||||
| CAZyme ID | MGYG000001375_02110 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2292291; End: 2294225 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 213 | 461 | 4.2e-91 | 0.9881422924901185 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 2.92e-26 | 213 | 458 | 2 | 272 | Cellulase (glycosyl hydrolase family 5). |
| cd14256 | Dockerin_I | 1.69e-10 | 569 | 628 | 1 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| pfam00404 | Dockerin_1 | 6.34e-04 | 570 | 627 | 1 | 55 | Dockerin type I repeat. The dockerin repeat is the binding partner of the cohesin domain pfam00963. The cohesin-dockerin interaction is the crucial interaction for complex formation in the cellulosome. The dockerin repeats, each bearing homology to the EF-hand calcium-binding loop bind calcium. |
| cd14253 | Dockerin | 0.007 | 570 | 628 | 1 | 56 | Dockerin repeat domain. Dockerins are modules in the cellulosome complex that often anchor catalytic subunits by binding to cohesin domains of scaffolding proteins. Three types of dockerins and their corresponding cohesin have been described in the literature. This alignment models two consecutive dockerin repeats, the functional unit. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL18180.1 | 0.0 | 1 | 644 | 1 | 644 |
| QJC54217.1 | 9.05e-107 | 203 | 500 | 32 | 328 |
| QUI25581.1 | 1.12e-106 | 203 | 500 | 20 | 314 |
| QAA30979.1 | 7.18e-105 | 203 | 500 | 30 | 324 |
| QGG58625.1 | 1.58e-104 | 203 | 500 | 32 | 328 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2WHJ_A | 4.77e-98 | 203 | 502 | 2 | 298 | Understandinghow diverse mannanases recognise heterogeneous substrates [Salipaludibacillus agaradhaerens] |
| 1WKY_A | 1.48e-97 | 195 | 500 | 1 | 303 | Crystalstructure of alkaline mannanase from Bacillus sp. strain JAMB-602: catalytic domain and its Carbohydrate Binding Module [Bacillus sp. JAMB-602] |
| 3JUG_A | 3.49e-96 | 203 | 500 | 24 | 318 | Crystalstructure of endo-beta-1,4-mannanase from the alkaliphilic Bacillus sp. N16-5 [Bacillus sp. N16-5] |
| 2WHL_A | 6.85e-93 | 203 | 500 | 1 | 293 | Understandinghow diverse mannanases recognise heterogeneous substrates [Salipaludibacillus agaradhaerens] |
| 1BQC_A | 9.89e-69 | 203 | 500 | 3 | 301 | Beta-MannanaseFrom Thermomonospora Fusca [Thermobifida fusca],2MAN_A Mannotriose Complex Of Thermomonospora Fusca Beta-Mannanase [Thermobifida fusca],3MAN_A Mannohexaose Complex Of Thermomonospora Fusca Beta-mannanase [Thermobifida fusca] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| G1K3N4 | 2.61e-97 | 203 | 502 | 2 | 298 | Mannan endo-1,4-beta-mannosidase OS=Salipaludibacillus agaradhaerens OX=76935 PE=1 SV=1 |
| P51529 | 1.18e-72 | 203 | 500 | 39 | 335 | Mannan endo-1,4-beta-mannosidase OS=Streptomyces lividans OX=1916 GN=manA PE=1 SV=2 |
| B3PF24 | 4.38e-62 | 198 | 500 | 43 | 345 | Mannan endo-1,4-beta-mannosidase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=man5A PE=1 SV=1 |
| P22533 | 1.22e-56 | 207 | 466 | 43 | 300 | Beta-mannanase/endoglucanase A OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=manA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
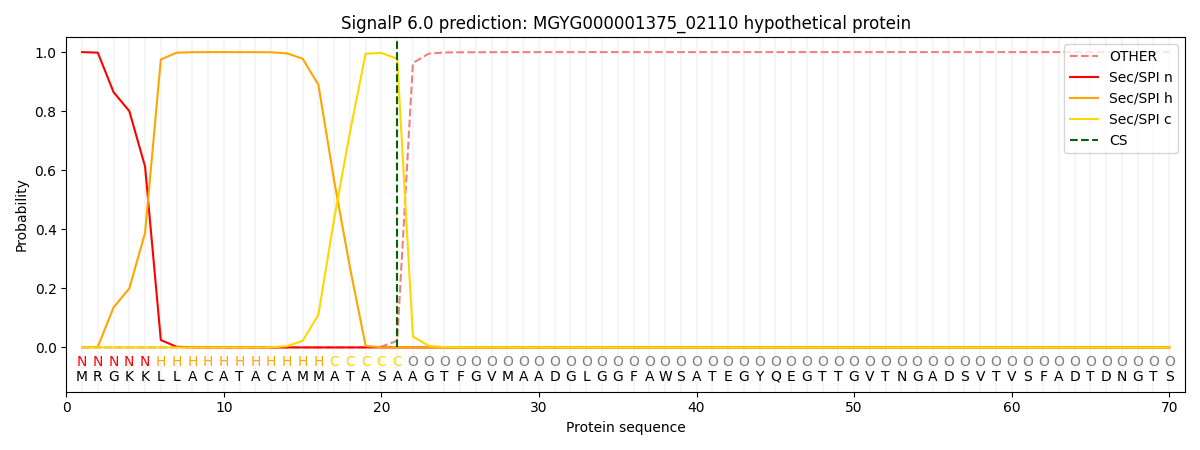
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000291 | 0.998918 | 0.000268 | 0.000177 | 0.000163 | 0.000154 |