You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001376_00088
Basic Information
help
| Species |
Dysgonomonas gadei
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Dysgonomonadaceae; Dysgonomonas; Dysgonomonas gadei
|
| CAZyme ID |
MGYG000001376_00088
|
| CAZy Family |
GH147 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001376 |
5166013 |
Isolate |
not provided |
not provided |
|
| Gene Location |
Start: 103029;
End: 105587
Strand: +
|
| Family |
Start |
End |
Evalue |
family coverage |
| GH147 |
18 |
833 |
0 |
0.9926470588235294 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG2730
|
BglC |
5.77e-04 |
82 |
249 |
83 |
240 |
Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 4QP0_A
|
1.81e-06 |
86 |
203 |
57 |
176 |
CrystalStructure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei [Rhizomucor miehei] |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
Q9SG94
|
1.64e-06 |
114 |
224 |
112 |
227 |
Mannan endo-1,4-beta-mannosidase 3 OS=Arabidopsis thaliana OX=3702 GN=MAN3 PE=2 SV=1 |
This protein is predicted as SP
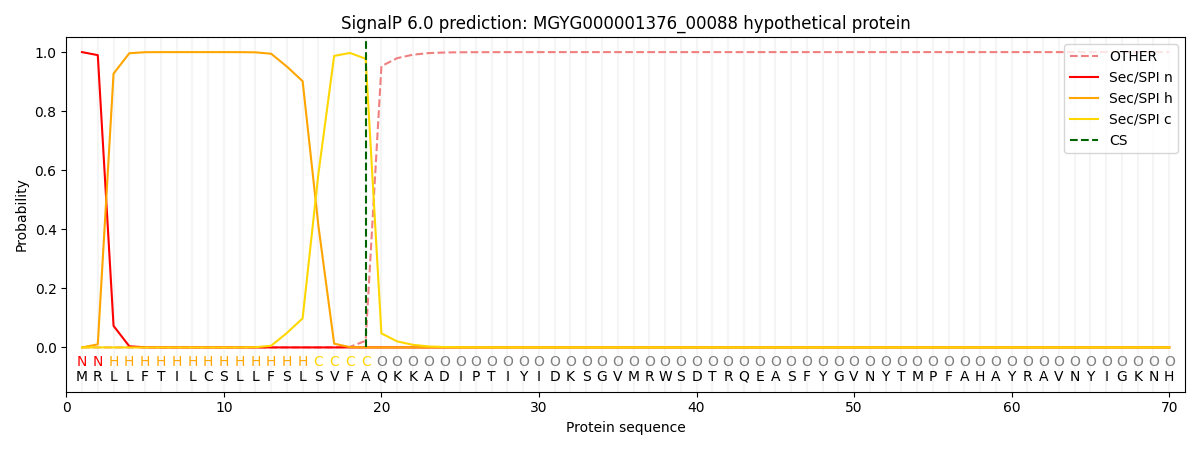
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000319
|
0.998901
|
0.000281
|
0.000165
|
0.000157
|
0.000150
|
There is no transmembrane helices in MGYG000001376_00088.

