You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001376_01553
You are here: Home > Sequence: MGYG000001376_01553
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
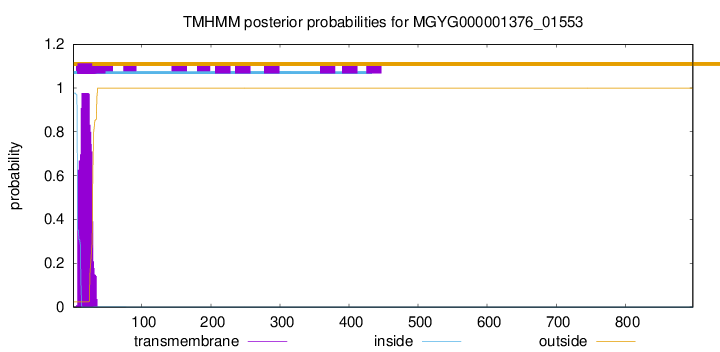
TMHMM annotations
Basic Information help
| Species | Dysgonomonas gadei | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Dysgonomonadaceae; Dysgonomonas; Dysgonomonas gadei | |||||||||||
| CAZyme ID | MGYG000001376_01553 | |||||||||||
| CAZy Family | GH11 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 231547; End: 234243 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH11 | 674 | 863 | 8e-69 | 0.9830508474576272 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00457 | Glyco_hydro_11 | 1.33e-71 | 673 | 862 | 1 | 175 | Glycosyl hydrolases family 11. |
| pfam03629 | SASA | 1.60e-14 | 407 | 509 | 98 | 200 | Carbohydrate esterase, sialic acid-specific acetylesterase. The catalytic triad of this esterase enzyme comprises residues Ser127, His403 and Asp391 in UniProtKB:P70665. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CCO21319.1 | 0.0 | 20 | 865 | 14 | 832 |
| CCO21108.1 | 0.0 | 20 | 865 | 14 | 832 |
| CCO21289.1 | 0.0 | 20 | 865 | 14 | 832 |
| CCO21210.1 | 0.0 | 20 | 865 | 14 | 832 |
| CCO21153.1 | 0.0 | 20 | 865 | 14 | 832 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7KMM_A | 1.31e-124 | 30 | 635 | 25 | 631 | ChainA, Sialic acid-specific 9-O-acetylesterase [Xanthomonas citri pv. citri str. 306],7KMM_B Chain B, Sialic acid-specific 9-O-acetylesterase [Xanthomonas citri pv. citri str. 306] |
| 2F6B_A | 5.25e-94 | 662 | 870 | 1 | 201 | Structuraland active site modification studies implicate Glu, Trp and Arg in the activity of xylanase from alkalophilic Bacillus sp. (NCL 87-6-10). [Bacillus],2F6B_B Structural and active site modification studies implicate Glu, Trp and Arg in the activity of xylanase from alkalophilic Bacillus sp. (NCL 87-6-10). [Bacillus] |
| 1H4G_A | 2.92e-93 | 663 | 870 | 2 | 201 | Oligosaccharide-bindingto family 11 xylanases: both covalent intermediate and mutant-product complexes display 2,5B conformations at the active-centre [Salipaludibacillus agaradhaerens],1H4G_B Oligosaccharide-binding to family 11 xylanases: both covalent intermediate and mutant-product complexes display 2,5B conformations at the active-centre [Salipaludibacillus agaradhaerens],1QH6_A CATALYSIS AND SPECIFICITY IN ENZYMATIC GLYCOSIDE HYDROLASES: A 2,5B CONFORMATION FOR THE GLYCOSYL-ENZYME INTERMIDIATE REVEALED BY THE STRUCTURE OF THE BACILLUS AGARADHAERENS FAMILY 11 XYLANASE [Salipaludibacillus agaradhaerens],1QH6_B CATALYSIS AND SPECIFICITY IN ENZYMATIC GLYCOSIDE HYDROLASES: A 2,5B CONFORMATION FOR THE GLYCOSYL-ENZYME INTERMIDIATE REVEALED BY THE STRUCTURE OF THE BACILLUS AGARADHAERENS FAMILY 11 XYLANASE [Salipaludibacillus agaradhaerens],1QH7_A CATALYSIS AND SPECIFICITY IN ENZYMATIC GLYCOSIDE HYDROLASES: A 2,5B CONFORMATION FOR THE GLYCOSYL-ENZYME INTERMIDIATE REVEALED BY THE STRUCTURE OF THE BACILLUS AGARADHAERENS FAMILY 11 XYLANASE [Salipaludibacillus agaradhaerens],1QH7_B CATALYSIS AND SPECIFICITY IN ENZYMATIC GLYCOSIDE HYDROLASES: A 2,5B CONFORMATION FOR THE GLYCOSYL-ENZYME INTERMIDIATE REVEALED BY THE STRUCTURE OF THE BACILLUS AGARADHAERENS FAMILY 11 XYLANASE [Salipaludibacillus agaradhaerens] |
| 1H4H_A | 8.55e-93 | 663 | 877 | 2 | 208 | Oligosaccharide-bindingto family 11 xylanases: both covalent intermediate and mutant-product complexes display 2,5B conformations at the active-centre [Salipaludibacillus agaradhaerens],1H4H_B Oligosaccharide-binding to family 11 xylanases: both covalent intermediate and mutant-product complexes display 2,5B conformations at the active-centre [Salipaludibacillus agaradhaerens],1H4H_C Oligosaccharide-binding to family 11 xylanases: both covalent intermediate and mutant-product complexes display 2,5B conformations at the active-centre [Salipaludibacillus agaradhaerens],1H4H_D Oligosaccharide-binding to family 11 xylanases: both covalent intermediate and mutant-product complexes display 2,5B conformations at the active-centre [Salipaludibacillus agaradhaerens] |
| 6KKA_A | 4.62e-90 | 664 | 871 | 2 | 201 | XylanaseJ mutant from Bacillus sp. 41M-1 [Bacillus sp. 41M-1],6KKA_B Xylanase J mutant from Bacillus sp. 41M-1 [Bacillus sp. 41M-1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8GJ44 | 2.26e-85 | 653 | 871 | 23 | 232 | Endo-1,4-beta-xylanase A OS=Thermoclostridium stercorarium OX=1510 GN=xynA PE=1 SV=2 |
| P17137 | 2.48e-85 | 658 | 871 | 57 | 260 | Endo-1,4-beta-xylanase OS=Clostridium saccharobutylicum OX=169679 GN=xynB PE=3 SV=1 |
| P83513 | 3.54e-83 | 661 | 878 | 16 | 223 | Bifunctional xylanase/deacetylase OS=Pseudobutyrivibrio xylanivorans OX=185007 GN=xyn11A PE=1 SV=2 |
| P33558 | 1.31e-82 | 653 | 871 | 23 | 233 | Endo-1,4-beta-xylanase A OS=Thermoclostridium stercorarium OX=1510 GN=xynA PE=1 SV=2 |
| P00694 | 7.03e-79 | 656 | 872 | 20 | 228 | Endo-1,4-beta-xylanase A OS=Bacillus pumilus OX=1408 GN=xynA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
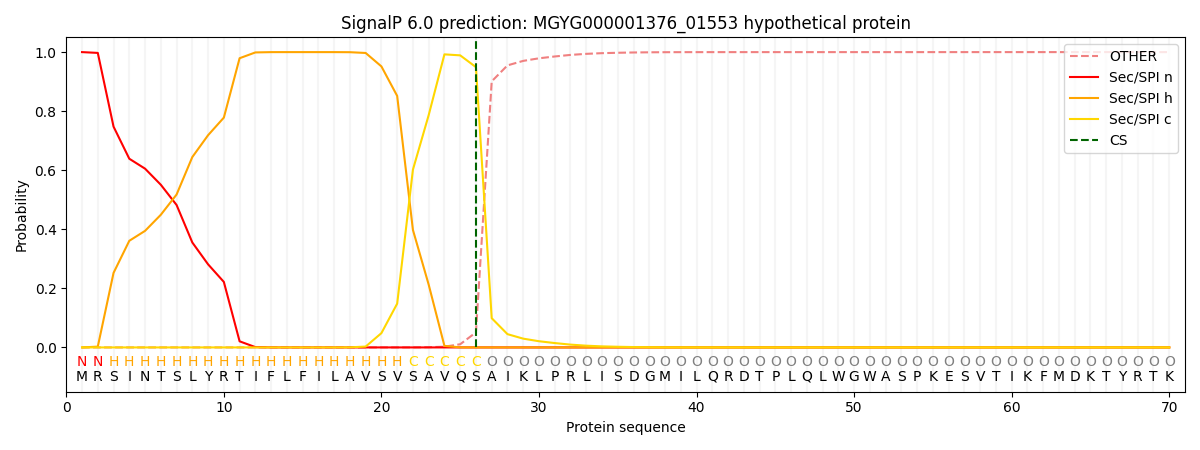
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000448 | 0.998914 | 0.000168 | 0.000153 | 0.000144 | 0.000142 |