You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001377_00769
You are here: Home > Sequence: MGYG000001377_00769
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Dysgonomonas mossii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Dysgonomonadaceae; Dysgonomonas; Dysgonomonas mossii | |||||||||||
| CAZyme ID | MGYG000001377_00769 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 45769; End: 47658 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 190 | 494 | 8.1e-44 | 0.9836065573770492 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRQ60219.1 | 2.16e-172 | 165 | 525 | 1 | 364 |
| QQT46979.1 | 2.65e-172 | 168 | 525 | 39 | 399 |
| QQT60504.1 | 1.51e-171 | 168 | 525 | 39 | 399 |
| APU67657.1 | 1.33e-164 | 171 | 562 | 42 | 433 |
| CBL16471.1 | 1.76e-56 | 179 | 528 | 33 | 364 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3VUP_A | 2.28e-12 | 181 | 493 | 4 | 322 | Beta-1,4-mannanasefrom the common sea hare Aplysia kurodai [Aplysia kurodai],3VUP_B Beta-1,4-mannanase from the common sea hare Aplysia kurodai [Aplysia kurodai] |
| 2C0H_A | 4.13e-11 | 179 | 499 | 5 | 332 | ChainA, MANNAN ENDO-1,4-BETA-MANNOSIDASE [Mytilus edulis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8WPJ2 | 2.46e-10 | 179 | 499 | 19 | 346 | Mannan endo-1,4-beta-mannosidase OS=Mytilus edulis OX=6550 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
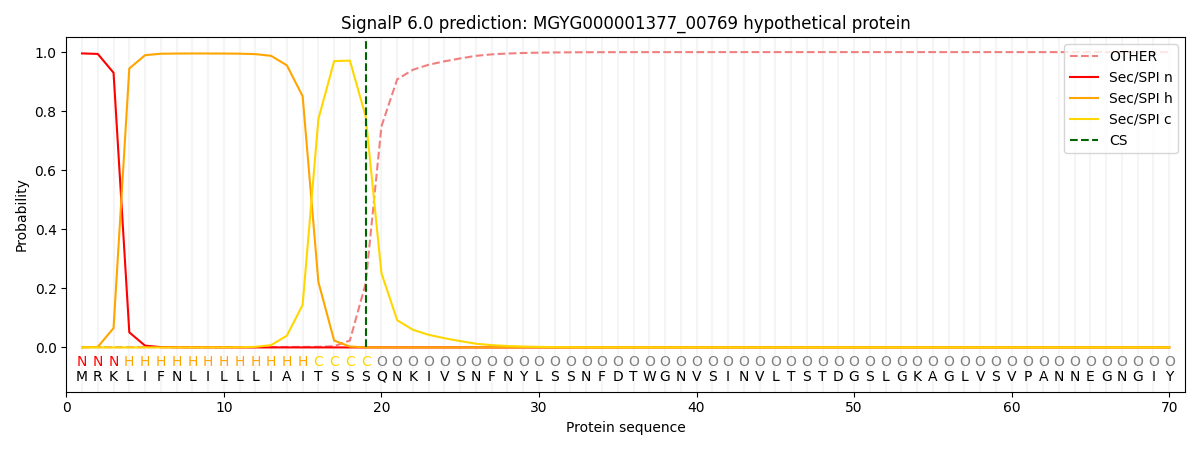
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002459 | 0.992155 | 0.004618 | 0.000236 | 0.000242 | 0.000262 |
