You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001377_01750
You are here: Home > Sequence: MGYG000001377_01750
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Dysgonomonas mossii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Dysgonomonadaceae; Dysgonomonas; Dysgonomonas mossii | |||||||||||
| CAZyme ID | MGYG000001377_01750 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 390772; End: 393597 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 27 | 740 | 1.4e-50 | 0.7513297872340425 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 8.18e-20 | 30 | 633 | 10 | 637 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 8.18e-09 | 30 | 342 | 10 | 350 | beta-D-glucuronidase; Provisional |
| pfam00703 | Glyco_hydro_2 | 3.43e-07 | 186 | 290 | 3 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT92878.1 | 0.0 | 30 | 940 | 28 | 934 |
| ALJ61552.1 | 0.0 | 30 | 940 | 28 | 934 |
| QUT34134.1 | 0.0 | 10 | 940 | 5 | 934 |
| QBJ17536.1 | 0.0 | 10 | 940 | 5 | 934 |
| QUU00164.1 | 0.0 | 10 | 940 | 5 | 934 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4JKM_A | 6.69e-08 | 25 | 417 | 8 | 419 | CrystalStructure of Clostridium perfringens beta-glucuronidase [Clostridium perfringens str. 13],4JKM_B Crystal Structure of Clostridium perfringens beta-glucuronidase [Clostridium perfringens str. 13],6CXS_A Crystal Structure of Clostridium perfringens beta-glucuronidase bound with a novel, potent inhibitor 4-(8-(piperazin-1-yl)-1,2,3,4-tetrahydro-[1,2,3]triazino[4',5':4,5]thieno[2,3-c]isoquinolin-5-yl)morpholine [Clostridium perfringens str. 13],6CXS_B Crystal Structure of Clostridium perfringens beta-glucuronidase bound with a novel, potent inhibitor 4-(8-(piperazin-1-yl)-1,2,3,4-tetrahydro-[1,2,3]triazino[4',5':4,5]thieno[2,3-c]isoquinolin-5-yl)morpholine [Clostridium perfringens str. 13] |
| 6D89_A | 4.07e-06 | 69 | 413 | 90 | 413 | Bacteroidesuniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis],6D89_B Bacteroides uniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis],6D89_C Bacteroides uniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis],6D89_D Bacteroides uniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis] |
| 6D1N_A | 4.10e-06 | 69 | 413 | 98 | 421 | Apostructure of Bacteroides uniformis Beta-glucuronidase 1 [Bacteroides uniformis],6D1N_B Apo structure of Bacteroides uniformis Beta-glucuronidase 1 [Bacteroides uniformis],6D41_A Bacteriodes uniformis beta-glucuronidase 1 bound to D-glucaro-1,5-lactone [Bacteroides uniformis],6D41_B Bacteriodes uniformis beta-glucuronidase 1 bound to D-glucaro-1,5-lactone [Bacteroides uniformis],6D6W_A Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D6W_B Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D6W_C Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D6W_D Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D7F_A Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_B Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_C Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_D Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_E Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_F Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
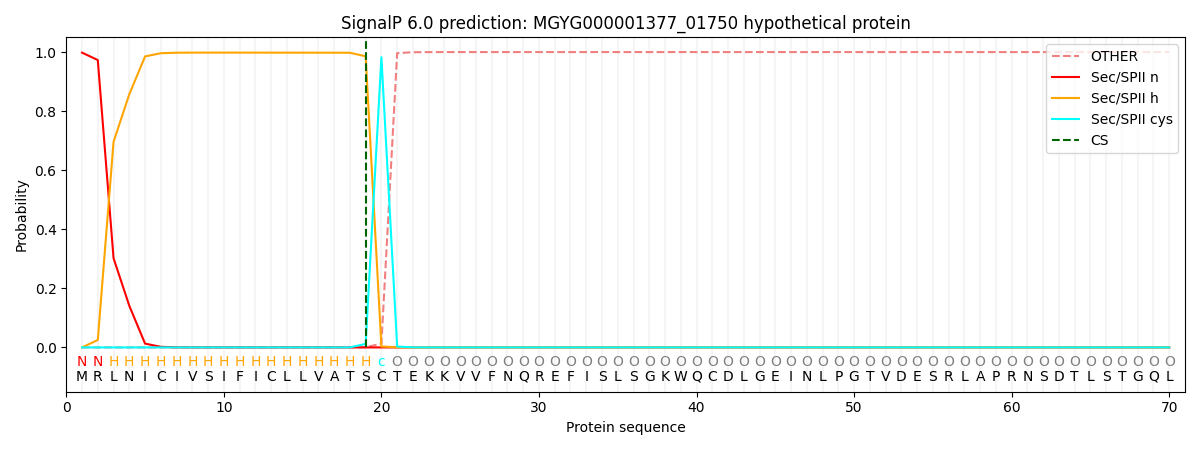
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000002 | 0.001720 | 0.998319 | 0.000001 | 0.000001 | 0.000001 |
