You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001378_02823
You are here: Home > Sequence: MGYG000001378_02823
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
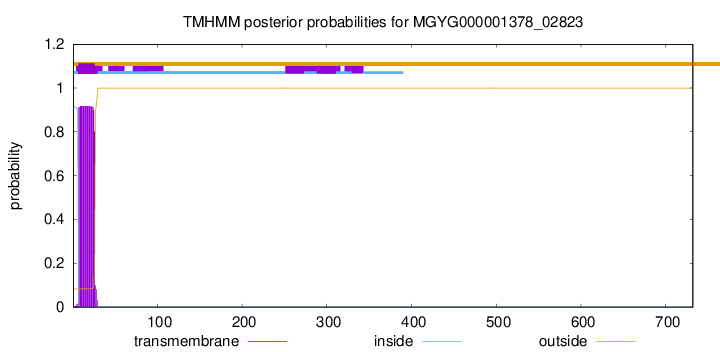
TMHMM annotations
Basic Information help
| Species | Bacteroides ovatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides ovatus | |||||||||||
| CAZyme ID | MGYG000001378_02823 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 321797; End: 323995 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 461 | 722 | 4.5e-46 | 0.6765676567656765 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 9.13e-34 | 465 | 723 | 100 | 310 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 2.44e-32 | 463 | 718 | 56 | 260 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.36e-22 | 463 | 718 | 121 | 334 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 5.67e-09 | 58 | 135 | 7 | 85 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 2.44e-07 | 101 | 149 | 8 | 56 | Glycosyl hydrolase family 10. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGT73069.1 | 0.0 | 1 | 732 | 1 | 732 |
| QUR44907.1 | 0.0 | 1 | 732 | 1 | 732 |
| QDM11106.1 | 0.0 | 1 | 730 | 1 | 730 |
| QIU93949.1 | 0.0 | 1 | 732 | 1 | 732 |
| QRQ48288.1 | 2.36e-298 | 2 | 725 | 9 | 716 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1I1W_A | 3.76e-15 | 465 | 726 | 104 | 302 | 0.89AUltra high resolution structure of a Thermostable Xylanase from Thermoascus Aurantiacus [Thermoascus aurantiacus],1I1X_A 1.11 A ATOMIC RESOLUTION STRUCTURE OF A THERMOSTABLE XYLANASE FROM THERMOASCUS AURANTIACUS [Thermoascus aurantiacus] |
| 6LPS_A | 6.46e-15 | 453 | 723 | 111 | 351 | ChainA, Beta-xylanase [Halalkalibacterium halodurans] |
| 2BNJ_A | 9.14e-15 | 465 | 726 | 104 | 302 | Thexylanase TA from Thermoascus aurantiacus utilizes arabinose decorations of xylan as significant substrate specificity determinants. [Thermoascus aurantiacus] |
| 7CPK_A | 1.26e-14 | 453 | 723 | 112 | 352 | XylanaseR from Bacillus sp. TAR-1 [Bacillus sp. TAR1] |
| 7CPL_A | 1.26e-14 | 453 | 723 | 112 | 352 | XylanaseR from Bacillus sp. TAR-1 [Bacillus sp. TAR1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O94163 | 3.51e-15 | 462 | 725 | 125 | 327 | Endo-1,4-beta-xylanase F1 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=xynF1 PE=1 SV=1 |
| Q96VB6 | 8.52e-14 | 465 | 725 | 126 | 323 | Endo-1,4-beta-xylanase F3 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=xynF3 PE=1 SV=1 |
| P07528 | 9.13e-14 | 453 | 723 | 156 | 396 | Endo-1,4-beta-xylanase A OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=xynA PE=1 SV=1 |
| P23360 | 2.19e-13 | 465 | 726 | 130 | 328 | Endo-1,4-beta-xylanase OS=Thermoascus aurantiacus OX=5087 GN=XYNA PE=1 SV=4 |
| Q9HEZ1 | 1.29e-12 | 465 | 727 | 195 | 408 | Endo-1,4-beta-xylanase A OS=Phanerodontia chrysosporium OX=2822231 GN=xynA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
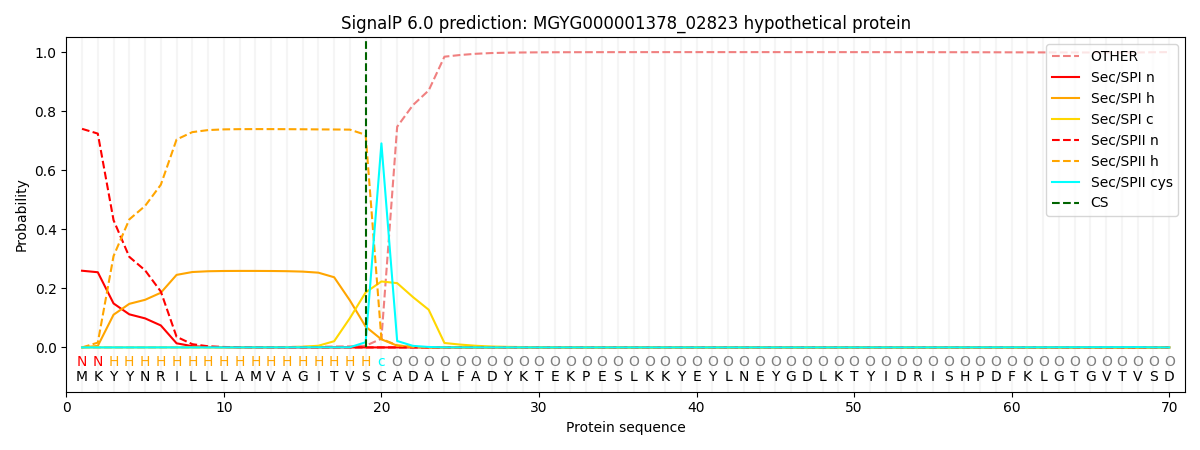
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000527 | 0.254414 | 0.744675 | 0.000122 | 0.000125 | 0.000125 |