You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001378_02977
You are here: Home > Sequence: MGYG000001378_02977
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides ovatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides ovatus | |||||||||||
| CAZyme ID | MGYG000001378_02977 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 95844; End: 97472 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 140 | 462 | 1.1e-57 | 0.9861111111111112 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 1.81e-27 | 121 | 409 | 58 | 360 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PLN02432 | PLN02432 | 9.13e-26 | 142 | 467 | 15 | 291 | putative pectinesterase |
| PLN02665 | PLN02665 | 1.78e-25 | 142 | 467 | 72 | 359 | pectinesterase family protein |
| PLN02773 | PLN02773 | 1.04e-21 | 142 | 407 | 9 | 246 | pectinesterase |
| PRK10531 | PRK10531 | 2.03e-21 | 139 | 384 | 81 | 346 | putative acyl-CoA thioester hydrolase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDM11674.1 | 0.0 | 1 | 542 | 1 | 542 |
| QGT73504.1 | 0.0 | 1 | 542 | 1 | 542 |
| QUT78075.1 | 0.0 | 1 | 542 | 1 | 542 |
| QNL41085.1 | 0.0 | 1 | 542 | 1 | 542 |
| QRQ55768.1 | 0.0 | 1 | 542 | 1 | 542 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4PMH_A | 8.09e-15 | 146 | 375 | 45 | 277 | Thestructure of rice weevil pectin methyl esterase [Sitophilus oryzae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q3E9D3 | 8.71e-17 | 136 | 434 | 25 | 298 | Probable pectinesterase 55 OS=Arabidopsis thaliana OX=3702 GN=PME55 PE=2 SV=1 |
| Q9LY18 | 3.96e-16 | 141 | 407 | 68 | 299 | Probable pectinesterase 49 OS=Arabidopsis thaliana OX=3702 GN=PME49 PE=2 SV=1 |
| A1DBT4 | 5.13e-15 | 130 | 421 | 24 | 285 | Probable pectinesterase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=pmeA PE=3 SV=1 |
| Q9LSP1 | 8.47e-15 | 141 | 417 | 47 | 293 | Probable pectinesterase 67 OS=Arabidopsis thaliana OX=3702 GN=PME67 PE=2 SV=1 |
| E7CIP7 | 1.17e-14 | 146 | 375 | 61 | 293 | Pectinesterase OS=Sitophilus oryzae OX=7048 GN=CE8-1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
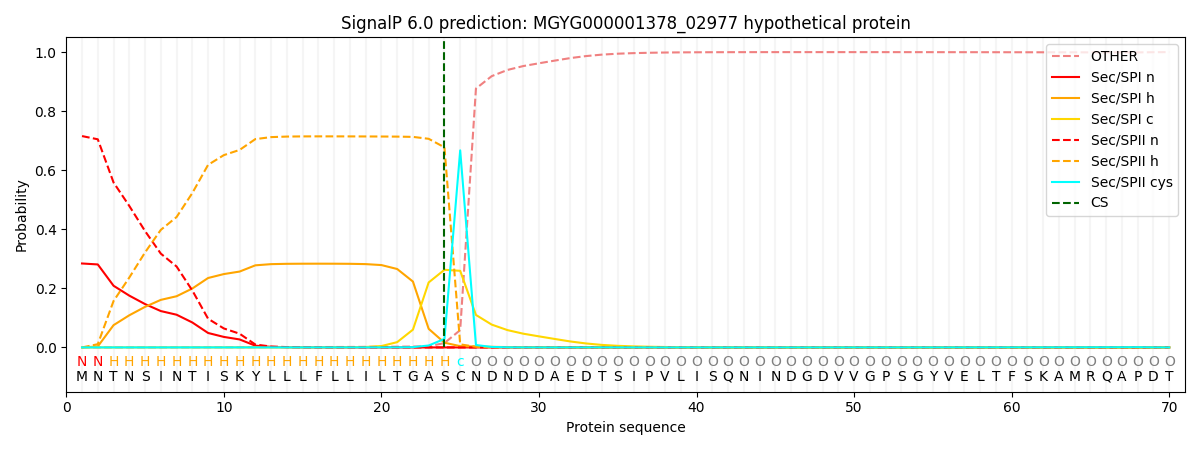
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000428 | 0.279250 | 0.720004 | 0.000109 | 0.000117 | 0.000097 |
