You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001378_03565
Basic Information
help
| Species |
Bacteroides ovatus
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides ovatus
|
| CAZyme ID |
MGYG000001378_03565
|
| CAZy Family |
PL27 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 694 |
|
80127.54 |
8.3077 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001378 |
6545242 |
Isolate |
not provided |
not provided |
|
| Gene Location |
Start: 411986;
End: 414070
Strand: +
|
| Family |
Start |
End |
Evalue |
family coverage |
| PL27 |
64 |
687 |
3.5e-266 |
0.9967213114754099 |
MGYG000001378_03565 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 5NO8_A
|
0.0 |
21 |
693 |
21 |
694 |
PolysaccharideLyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838],5NO8_B Polysaccharide Lyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838] |
| 5NOK_A
|
0.0 |
21 |
693 |
21 |
694 |
PolysaccharideLyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838],5NOK_B Polysaccharide Lyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838] |
This protein is predicted as SP
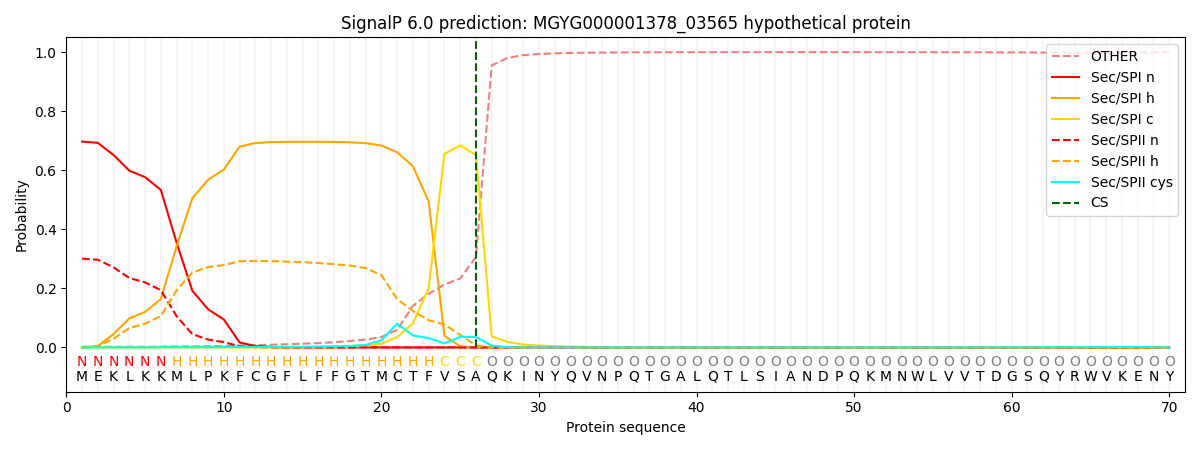
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.003702
|
0.685584
|
0.309716
|
0.000386
|
0.000285
|
0.000298
|
There is no transmembrane helices in MGYG000001378_03565.

