You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001383_00571
You are here: Home > Sequence: MGYG000001383_00571
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
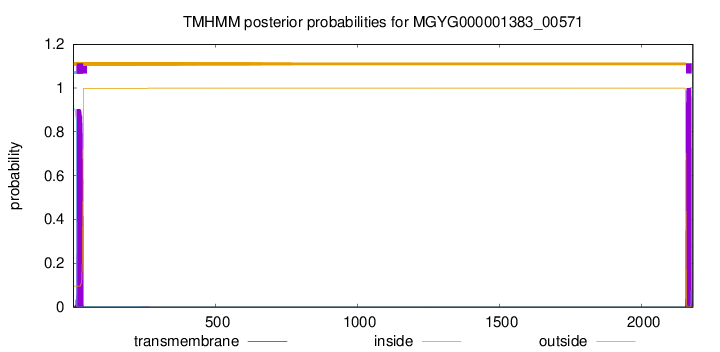
TMHMM annotations
Basic Information help
| Species | Lactobacillus iners | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Lactobacillus; Lactobacillus iners | |||||||||||
| CAZyme ID | MGYG000001383_00571 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 611665; End: 618210 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 49 | 184 | 7.3e-27 | 0.9925373134328358 |
| CBM32 | 799 | 924 | 5.2e-17 | 0.8870967741935484 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PHA03247 | PHA03247 | 2.42e-31 | 1794 | 2117 | 2702 | 2997 | large tegument protein UL36; Provisional |
| PHA03247 | PHA03247 | 2.59e-31 | 1793 | 2110 | 2576 | 2930 | large tegument protein UL36; Provisional |
| PTZ00449 | PTZ00449 | 5.25e-31 | 1798 | 2088 | 531 | 814 | 104 kDa microneme/rhoptry antigen; Provisional |
| PTZ00449 | PTZ00449 | 5.92e-31 | 1784 | 2110 | 565 | 929 | 104 kDa microneme/rhoptry antigen; Provisional |
| NF033839 | PspC_subgroup_2 | 4.30e-29 | 1780 | 2106 | 249 | 499 | pneumococcal surface protein PspC, LPXTG-anchored form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. The other form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIH26888.1 | 0.0 | 1 | 1810 | 1 | 1810 |
| QIH28268.1 | 0.0 | 1 | 1810 | 1 | 1810 |
| QIH25566.1 | 0.0 | 1 | 1810 | 1 | 1810 |
| QIH23015.1 | 0.0 | 1 | 1810 | 1 | 1810 |
| QIH24239.1 | 0.0 | 1 | 1810 | 1 | 1810 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7JS4_A | 8.87e-13 | 26 | 208 | 578 | 772 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 4A41_A | 7.68e-11 | 791 | 924 | 28 | 154 | CpGH89CBM32-5,from Clostridium perfringens, in complex with galactose [Clostridium perfringens],4A44_A CpGH89CBM32-5, from Clostridium perfringens, in complex with the Tn Antigen [Clostridium perfringens],4A45_A CpGH89CBM32-5, from Clostridium perfringens, in complex with GalNAc- beta-1,3-galactose [Clostridium perfringens],4AAX_A CpGH89CBM32-5, from Clostridium perfringens, in complex with N- acetylgalactosamine [Clostridium perfringens] |
| 2J7M_A | 6.84e-10 | 792 | 924 | 7 | 142 | Characterizationof a Family 32 CBM [Clostridium perfringens] |
| 2J1A_A | 7.00e-10 | 792 | 924 | 8 | 143 | Structureof CBM32 from Clostridium perfringens beta-N- acetylhexosaminidase GH84C in complex with galactose [Clostridium perfringens ATCC 13124],2J1E_A High Resolution Crystal Structure of CBM32 from a N-acetyl-beta- hexosaminidase in complex with lacNAc [Clostridium perfringens ATCC 13124] |
| 2V5D_A | 4.52e-09 | 787 | 924 | 590 | 730 | Structureof a Family 84 Glycoside Hydrolase and a Family 32 Carbohydrate-Binding Module in Tandem from Clostridium perfringens. [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0TR53 | 2.83e-08 | 787 | 924 | 620 | 760 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| Q8XL08 | 2.83e-08 | 787 | 924 | 620 | 760 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
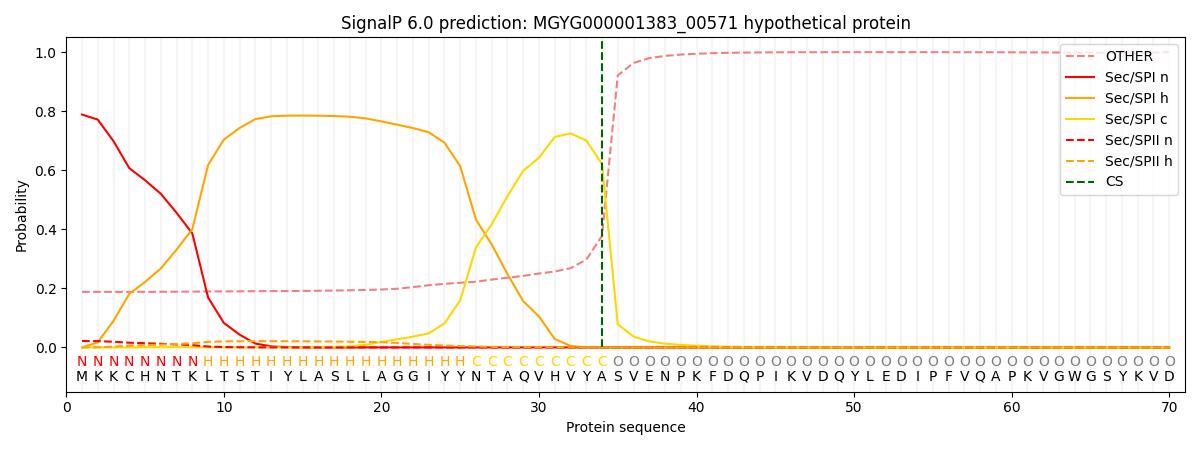
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.202299 | 0.771497 | 0.022604 | 0.002664 | 0.000491 | 0.000411 |