You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001384_01695
Basic Information
help
| Species |
Ralstonia pickettii
|
| Lineage |
Bacteria; Proteobacteria; Gammaproteobacteria; Burkholderiales; Burkholderiaceae; Ralstonia; Ralstonia pickettii
|
| CAZyme ID |
MGYG000001384_01695
|
| CAZy Family |
GT83 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001384 |
5243975 |
Isolate |
not provided |
not provided |
|
| Gene Location |
Start: 1784966;
End: 1786990
Strand: +
|
No EC number prediction in MGYG000001384_01695.
| Family |
Start |
End |
Evalue |
family coverage |
| GT83 |
34 |
576 |
1.4e-43 |
0.8703703703703703 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG1807
|
ArnT |
1.95e-43 |
33 |
648 |
3 |
535 |
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231
|
PMT_2 |
0.002 |
94 |
300 |
2 |
160 |
Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
A8GDR9
|
1.37e-08 |
39 |
324 |
8 |
248 |
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Serratia proteamaculans (strain 568) OX=399741 GN=arnT PE=3 SV=1 |
|
A1JPM4
|
7.94e-06 |
58 |
324 |
27 |
248 |
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Yersinia enterocolitica serotype O:8 / biotype 1B (strain NCTC 13174 / 8081) OX=393305 GN=arnT PE=3 SV=1 |
This protein is predicted as OTHER
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.999969
|
0.000044
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
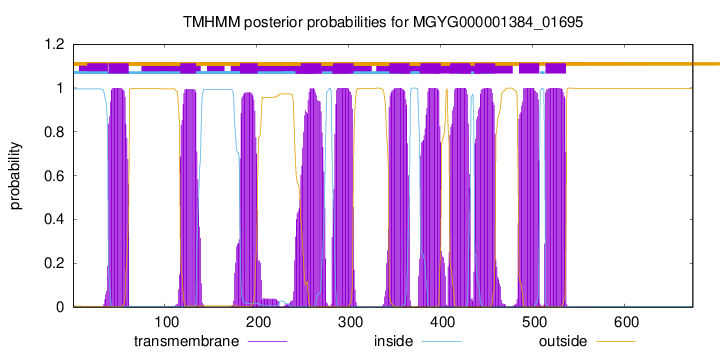
| start |
end |
| 39 |
61 |
| 117 |
134 |
| 182 |
201 |
| 248 |
270 |
| 283 |
305 |
| 344 |
366 |
| 378 |
400 |
| 410 |
432 |
| 437 |
459 |
| 485 |
507 |
| 514 |
536 |


