You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001384_01967
You are here: Home > Sequence: MGYG000001384_01967
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
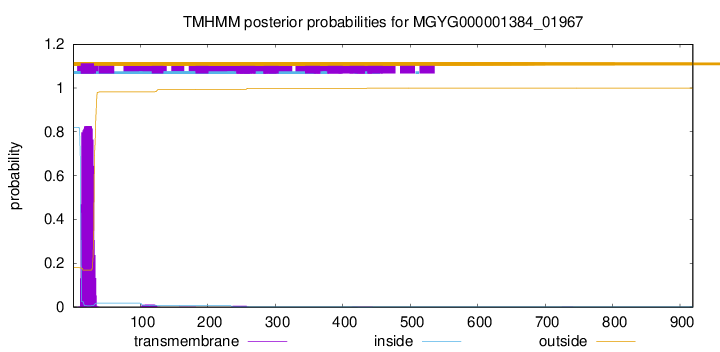
TMHMM annotations
Basic Information help
| Species | Ralstonia pickettii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Burkholderiales; Burkholderiaceae; Ralstonia; Ralstonia pickettii | |||||||||||
| CAZyme ID | MGYG000001384_01967 | |||||||||||
| CAZy Family | GH166 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2065706; End: 2068465 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH166 | 98 | 250 | 2.9e-23 | 0.7098214285714286 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd10922 | CE4_PelA_like_C | 8.21e-138 | 480 | 751 | 1 | 266 | C-terminal Putative NodB-like catalytic domain of PelA-like uncharacterized hypothetical proteins found in bacteria. This family is represented by a protein PelA of unknown function that is encoded by a gene in the pelA-G gene cluster for pellicle production and biofilm formation in Pseudomonas aeruginosa. PelA and most of the family members contain a domain of unknown function, DUF297, in the N-terminus and a C-terminal domain that shows high sequence similarity to the catalytic domain of the six-stranded barrel rhizobial NodB-like proteins, which remove N-linked or O-linked acetyl groups from cell wall polysaccharides and belong to the larger carbohydrate esterase 4 (CE4) superfamily. |
| COG3868 | COG3868 | 9.89e-78 | 15 | 320 | 1 | 306 | Predicted glycosyl hydrolase, GH114 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQK36622.1 | 0.0 | 1 | 919 | 1 | 919 |
| ANJ73984.1 | 0.0 | 1 | 917 | 1 | 912 |
| ANH73464.1 | 0.0 | 1 | 917 | 1 | 912 |
| AJW43534.1 | 0.0 | 1 | 917 | 1 | 923 |
| ANA33257.1 | 0.0 | 1 | 917 | 1 | 925 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5TCB_A | 2.20e-17 | 46 | 264 | 27 | 278 | Structureof the glycoside hydrolase domain of PelA from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1] |
| 5TSY_A | 2.97e-17 | 46 | 264 | 27 | 278 | Structureof the glycoside hydrolase domain of PelA variant E218A from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
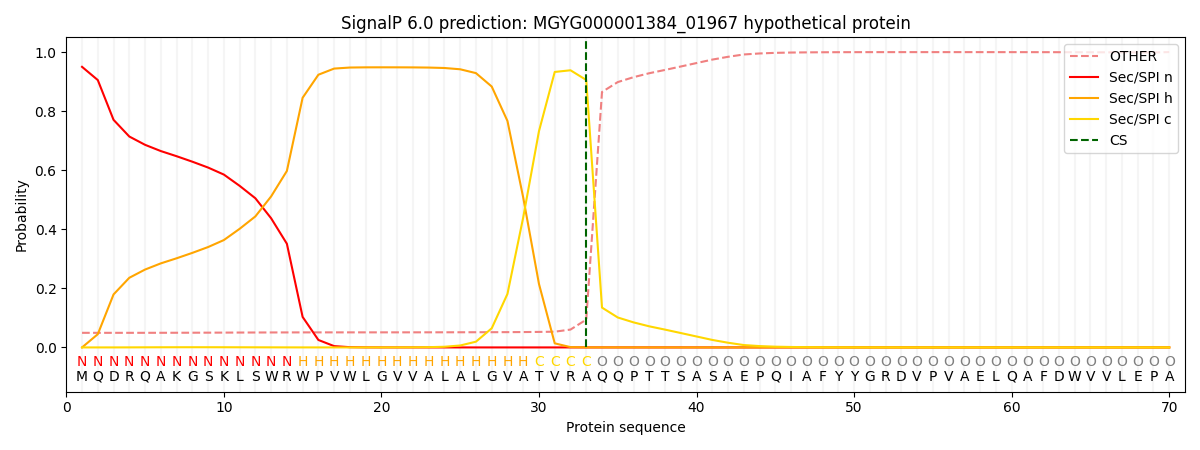
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.053290 | 0.944533 | 0.000957 | 0.000537 | 0.000338 | 0.000303 |