You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001384_02447
You are here: Home > Sequence: MGYG000001384_02447
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Ralstonia pickettii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Burkholderiales; Burkholderiaceae; Ralstonia; Ralstonia pickettii | |||||||||||
| CAZyme ID | MGYG000001384_02447 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | Protein TolB | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 405625; End: 407043 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 351 | 468 | 5.2e-27 | 0.9435483870967742 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0823 | TolB | 9.18e-38 | 5 | 470 | 6 | 425 | Periplasmic component of the Tol biopolymer transport system [Intracellular trafficking, secretion, and vesicular transport]. |
| pfam00754 | F5_F8_type_C | 7.59e-17 | 361 | 464 | 13 | 123 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 2.27e-08 | 385 | 452 | 46 | 127 | Substituted updates: Jan 31, 2002 |
| PRK00178 | tolB | 1.75e-05 | 90 | 263 | 206 | 384 | Tol-Pal system protein TolB. |
| PRK04922 | tolB | 0.001 | 46 | 146 | 217 | 308 | Tol-Pal system beta propeller repeat protein TolB. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQK37331.1 | 0.0 | 1 | 472 | 1 | 472 |
| ATG22258.1 | 4.57e-307 | 1 | 472 | 1 | 474 |
| ANA35075.1 | 9.22e-307 | 1 | 472 | 1 | 474 |
| QIF10083.1 | 7.57e-306 | 1 | 472 | 1 | 474 |
| AJW46734.1 | 7.57e-306 | 1 | 472 | 1 | 474 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2RV9_A | 6.05e-20 | 344 | 471 | 8 | 136 | Solutionstructure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 4ZXE_A | 6.21e-20 | 344 | 471 | 9 | 137 | X-raycrystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_B X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_C X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis] |
| 4ZY9_A | 1.17e-19 | 344 | 471 | 9 | 137 | X-raycrystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZY9_B X-ray crystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 2RVA_A | 1.75e-17 | 365 | 471 | 29 | 137 | Solutionstructure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 4ZZ5_A | 1.80e-17 | 365 | 471 | 30 | 138 | X-raycrystal structure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZZ5_B X-ray crystal structure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZZ8_A X-ray crystal structure of chitosan-binding module 2 in complex with chitotriose derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZZ8_B X-ray crystal structure of chitosan-binding module 2 in complex with chitotriose derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
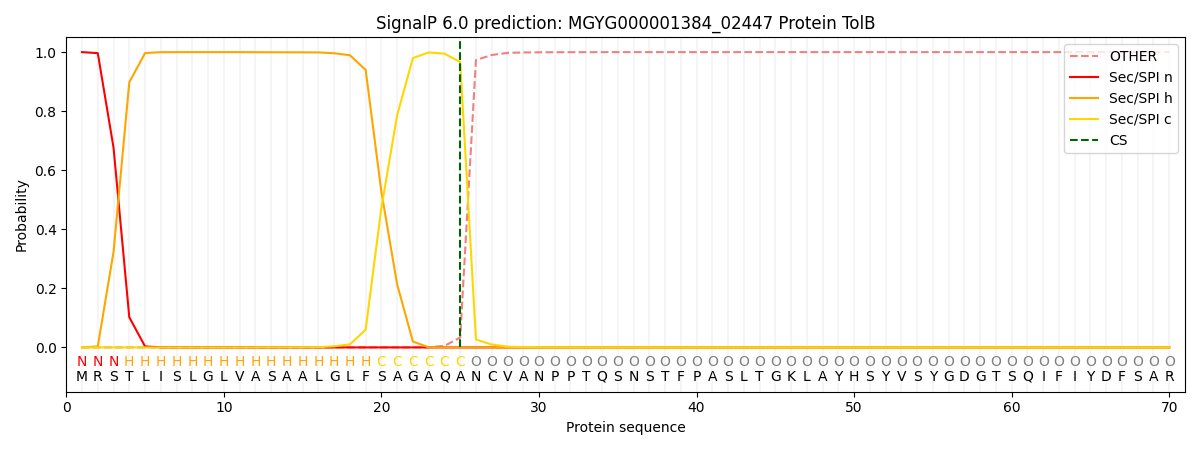
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000385 | 0.998829 | 0.000233 | 0.000195 | 0.000163 | 0.000153 |
